Aldehyde dehydrogenase: Difference between revisions
Jump to navigation
Jump to search
Michal Harel (talk | contribs) No edit summary |
Michal Harel (talk | contribs) No edit summary |
||
| (31 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
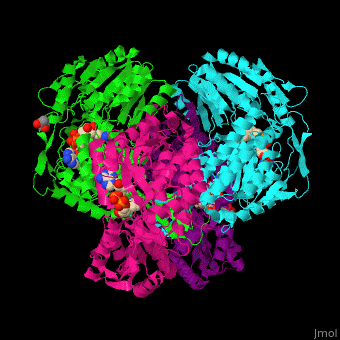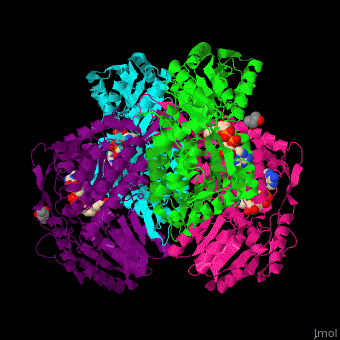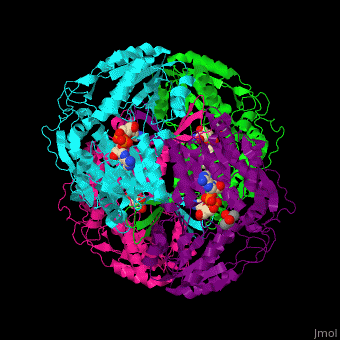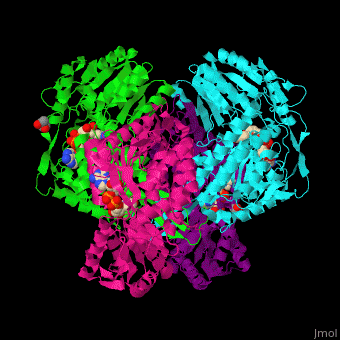
<StructureSection load='' size='450' side='right' scene='44/447036/Cv/2' caption='Aldehyde hydrogenase class 1 tetramer complex with NAD, dithiothreitol and dimercaptobutane-diol, [[1o9j]]'> | |||
== Function == | |||
'''Aldehyde dehydrogenase''' (ALDH) converts aldehydes to carboxylic acids while reducing NAD+ to NADH. In mammals there are 3 classes of ALDH and each contain constitutive and inducible forms.<br /> | |||
* '''ALDH class 1''' is cytosolic.<br /> | |||
* '''RALDH class 1A 1''' is retinal dehydrogenase which converts retinalaldehyde to retinoic acid.<br /> | |||
* '''RALDH class 1A 2''' is retinal dehydrogenase which converts retinal to retinoate.<br /> | |||
* '''RALDH class 1A 3''' is retinal dehydrogenase which funcrions in detoxification of aldehydes.<br /> | |||
* '''ALDH class 2''' is mitochondrial. <br /> | |||
* '''ALDH class 3''' is found in tumors, stomach and cornea. '''ALDH3A1''' is soluble and has substrate specificity to bulky aromatic aldehydes. '''ALDH3A2''' is a fatty ALDH (FALDH). FALDH was found to have an additional gatekeeper helix at the substrate funnel entrance that is shaping the enzymes substrate specificity. <ref>Keller, Markus A.; Zander, Ulrich; Fuchs, Julian E.; Kreutz, Christoph; Watschinger, Katrin et al. (2014). A gatekeeper helix determines the substrate specificity of Sjögren–Larsson Syndrome enzyme fatty aldehyde dehydrogenase. Nature Communications vol. 5.</ref><br /> | |||
* '''ALDH family 7 member A1''' is known as '''antiquitin''' and functions in the detoxification of aldehydes. <br /> | |||
* '''Glyceraldehyde-3-phophate (G3P)-ALDH''' is called GAPDH. GADPH catalyzes the reversible oxidative phosphorylation of glyceraldehyde-3-phosphate (G3P) to 1,3-bisphosphoglycerate in the presence of inorganic phosphate (Pi) and NAD. The aldehyde of G3P reacts with the cysteine-thiol to form a carboxylic acid in a high energy thioester form. The thioester is attacked by the inorganic phosphate and forms the acyl phosphate. GAPDH is part of the [[glycolysis]] pathway and [[Calvin cycle]]. GAPDH contains NAD-dependent and NADPH-dependent enzymes. For the complex of ALDH and nitroglycerine see [[NitroDur]]. | |||
* '''Indole-3-acetaldehyde dehydrogenase''' catalyzes the formation of indole-3-acetic acid from indole-3-acetaldehyde<ref>PMID:29293681</ref>.<br /> | |||
See also: | |||
*[[Pyrroline-5-carboxylate dehydrogenase]] | |||
*[[Succinate-semialdehyde dehydrogenase]] | |||
== Disease == | |||
ALDH inhibition is associate with Parkinson disease. Inhibitors can include pesticides. Mitochondrial ALDH2 deficiency causes accumulation of acetaldehyde in the blood following alcohol consumption resulting in the syndrome known as Alcohol flush syndrome. | |||
== Relevance == | |||
Buildup of toxic aldehydes causes cell damage and cardiac diseases. | |||
== Structural highlights == | |||
A cysteine-thiol at the active site of GAPDH plays a role in catalysis. | |||
*<scene name='44/447036/Cv/9'>Active site</scene>. Water molecules shown as red spheres. | |||
*<scene name='44/447036/Cv/10'>Dithiothreitol binding</scene>. | |||
*<scene name='44/447036/Cv/11'>Dimercaptobutane-diol binding</scene>. | |||
== 3D Structures of aldehyde dehydrogenase == | |||
[[Aldehyde dehydrogenase 3D structures]] | |||
</StructureSection> | |||
==References== | |||
{{reflist}} | |||
<references/> | |||
== | |||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Latest revision as of 11:44, 22 May 2024
FunctionAldehyde dehydrogenase (ALDH) converts aldehydes to carboxylic acids while reducing NAD+ to NADH. In mammals there are 3 classes of ALDH and each contain constitutive and inducible forms.
See also: DiseaseALDH inhibition is associate with Parkinson disease. Inhibitors can include pesticides. Mitochondrial ALDH2 deficiency causes accumulation of acetaldehyde in the blood following alcohol consumption resulting in the syndrome known as Alcohol flush syndrome. RelevanceBuildup of toxic aldehydes causes cell damage and cardiac diseases. Structural highlightsA cysteine-thiol at the active site of GAPDH plays a role in catalysis.
3D Structures of aldehyde dehydrogenaseAldehyde dehydrogenase 3D structures
|
| ||||||||||
ReferencesReferences
- ↑ Keller, Markus A.; Zander, Ulrich; Fuchs, Julian E.; Kreutz, Christoph; Watschinger, Katrin et al. (2014). A gatekeeper helix determines the substrate specificity of Sjögren–Larsson Syndrome enzyme fatty aldehyde dehydrogenase. Nature Communications vol. 5.
- ↑ McClerklin SA, Lee SG, Harper CP, Nwumeh R, Jez JM, Kunkel BN. Indole-3-acetaldehyde dehydrogenase-dependent auxin synthesis contributes to virulence of Pseudomonas syringae strain DC3000. PLoS Pathog. 2018 Jan 2;14(1):e1006811. doi: 10.1371/journal.ppat.1006811., eCollection 2018 Jan. PMID:29293681 doi:http://dx.doi.org/10.1371/journal.ppat.1006811