1fox: Difference between revisions
No edit summary |
No edit summary |
||
| (13 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
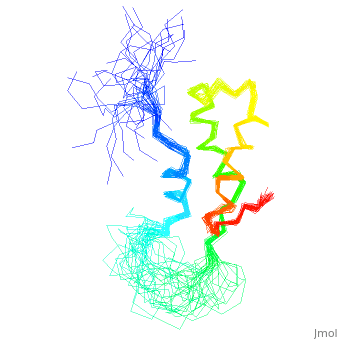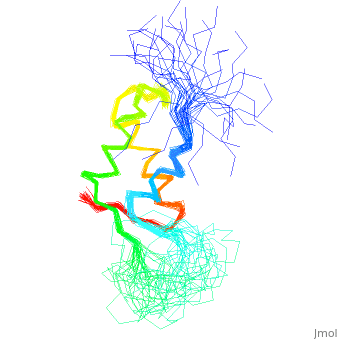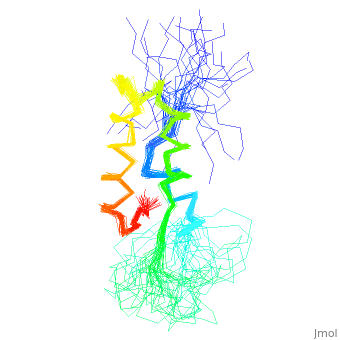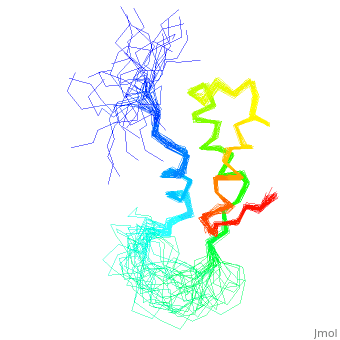
< | ==NMR STRUCTURE OF L11-C76, THE C-TERMINAL DOMAIN OF 50S RIBOSOMAL PROTEIN L11, 33 STRUCTURES== | ||
<StructureSection load='1fox' size='340' side='right'caption='[[1fox]]' scene=''> | |||
You may | == Structural highlights == | ||
<table><tr><td colspan='2'>[[1fox]] is a 1 chain structure with sequence from [https://en.wikipedia.org/wiki/Geobacillus_stearothermophilus Geobacillus stearothermophilus]. Full experimental information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1FOX OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=1FOX FirstGlance]. <br> | |||
</td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">Solution NMR</td></tr> | |||
- | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1fox FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1fox OCA], [https://pdbe.org/1fox PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1fox RCSB], [https://www.ebi.ac.uk/pdbsum/1fox PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1fox ProSAT]</span></td></tr> | ||
</table> | |||
== Function == | |||
[https://www.uniprot.org/uniprot/RL11_GEOSE RL11_GEOSE] Forms part of the ribosomal stalk which helps the ribosome interact with GTP-bound translation factors. | |||
== Evolutionary Conservation == | |||
[[Image:Consurf_key_small.gif|200px|right]] | |||
Check<jmol> | |||
<jmolCheckbox> | |||
<scriptWhenChecked>; select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/fo/1fox_consurf.spt"</scriptWhenChecked> | |||
<scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> | |||
<text>to colour the structure by Evolutionary Conservation</text> | |||
</jmolCheckbox> | |||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1fox ConSurf]. | |||
<div style="clear:both"></div> | |||
<div style="background-color:#fffaf0;"> | |||
== Publication Abstract from PubMed == | |||
The structure of the C-terminal RNA recognition domain of ribosomal protein L11 has been solved by heteronuclear three-dimensional nuclear magnetic resonance spectroscopy. Although the structure can be considered high resolution in the core, 15 residues between helix alpha 1 and strand beta 1 form an extended, unstructured loop. 15N transverse relaxation measurements suggest that the loop is moving on a picosecond-to-nanosecond time scale in the free protein but not in the protein bound to RNA. Chemical shifts differences between the free protein and the bound protein suggest that the loop as well as the C-terminal end of helix alpha 3 are involved in RNA binding. | |||
High resolution solution structure of ribosomal protein L11-C76, a helical protein with a flexible loop that becomes structured upon binding to RNA.,Markus MA, Hinck AP, Huang S, Draper DE, Torchia DA Nat Struct Biol. 1997 Jan;4(1):70-7. PMID:8989327<ref>PMID:8989327</ref> | |||
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | |||
</div> | |||
<div class="pdbe-citations 1fox" style="background-color:#fffaf0;"></div> | |||
==See Also== | |||
*[[Ribosomal protein L11 3D structures|Ribosomal protein L11 3D structures]] | |||
== References == | |||
<references/> | |||
__TOC__ | |||
</StructureSection> | |||
== | |||
== | |||
[[Category: Geobacillus stearothermophilus]] | [[Category: Geobacillus stearothermophilus]] | ||
[[Category: | [[Category: Large Structures]] | ||
[[Category: Draper | [[Category: Draper DE]] | ||
[[Category: Hinck | [[Category: Hinck AP]] | ||
[[Category: Huang | [[Category: Huang S]] | ||
[[Category: Markus | [[Category: Markus MA]] | ||
[[Category: Torchia | [[Category: Torchia DA]] | ||
Latest revision as of 11:29, 22 May 2024
NMR STRUCTURE OF L11-C76, THE C-TERMINAL DOMAIN OF 50S RIBOSOMAL PROTEIN L11, 33 STRUCTURESNMR STRUCTURE OF L11-C76, THE C-TERMINAL DOMAIN OF 50S RIBOSOMAL PROTEIN L11, 33 STRUCTURES
Structural highlights
FunctionRL11_GEOSE Forms part of the ribosomal stalk which helps the ribosome interact with GTP-bound translation factors. Evolutionary Conservation Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. Publication Abstract from PubMedThe structure of the C-terminal RNA recognition domain of ribosomal protein L11 has been solved by heteronuclear three-dimensional nuclear magnetic resonance spectroscopy. Although the structure can be considered high resolution in the core, 15 residues between helix alpha 1 and strand beta 1 form an extended, unstructured loop. 15N transverse relaxation measurements suggest that the loop is moving on a picosecond-to-nanosecond time scale in the free protein but not in the protein bound to RNA. Chemical shifts differences between the free protein and the bound protein suggest that the loop as well as the C-terminal end of helix alpha 3 are involved in RNA binding. High resolution solution structure of ribosomal protein L11-C76, a helical protein with a flexible loop that becomes structured upon binding to RNA.,Markus MA, Hinck AP, Huang S, Draper DE, Torchia DA Nat Struct Biol. 1997 Jan;4(1):70-7. PMID:8989327[1] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences |
| ||||||||||||||||