4zp3: Difference between revisions
New page: '''Unreleased structure''' The entry 4zp3 is ON HOLD Authors: Goetz, F., Roske, Y., Faelber, K., Zuehlke, K., Autenrieth, K., Kreuchwig, A., Krause, G., Herberg, F.W., Daumke, O., Heine... |
No edit summary |
||
| (6 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
==AKAP18:PKA-RIIalpha structure reveals crucial anchor points for recognition of regulatory subunits of PKA== | |||
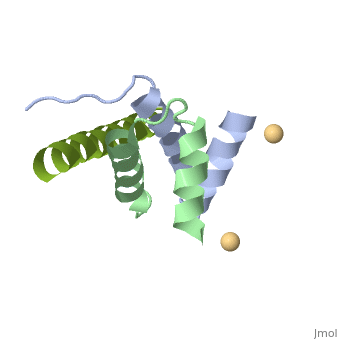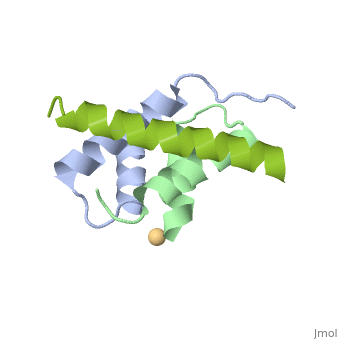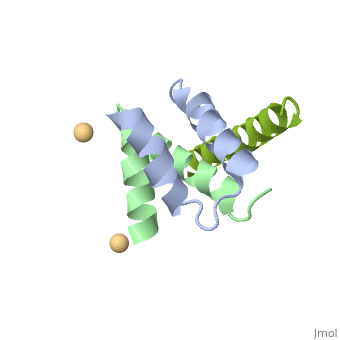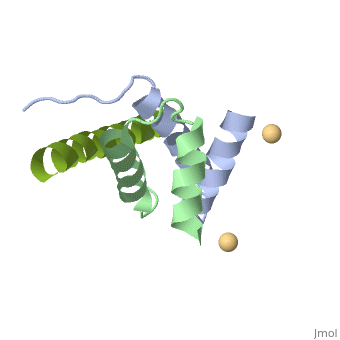
<StructureSection load='4zp3' size='340' side='right'caption='[[4zp3]], [[Resolution|resolution]] 2.63Å' scene=''> | |||
== Structural highlights == | |||
<table><tr><td colspan='2'>[[4zp3]] is a 18 chain structure with sequence from [https://en.wikipedia.org/wiki/Homo_sapiens Homo sapiens]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=4ZP3 OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=4ZP3 FirstGlance]. <br> | |||
</td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 2.63Å</td></tr> | |||
<tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=CD:CADMIUM+ION'>CD</scene></td></tr> | |||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=4zp3 FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=4zp3 OCA], [https://pdbe.org/4zp3 PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=4zp3 RCSB], [https://www.ebi.ac.uk/pdbsum/4zp3 PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=4zp3 ProSAT]</span></td></tr> | |||
</table> | |||
== Function == | |||
[https://www.uniprot.org/uniprot/AKA7G_HUMAN AKA7G_HUMAN] Probably targets cAMP-dependent protein kinase (PKA) to the cellular membrane or cytoskeletal structures. The membrane-associated form reduces epithelial sodium channel (ENaC) activity, whereas the free cytoplasmic form may negatively regulate ENaC channel feedback inhibition by intracellular sodium.<ref>PMID:10613906</ref> <ref>PMID:17244820</ref> | |||
<div style="background-color:#fffaf0;"> | |||
== Publication Abstract from PubMed == | |||
A-kinase anchoring proteins (AKAPs) interact with the dimerization/docking (D/D) domains of regulatory subunits of the ubiquitous protein kinase A (PKA). AKAPs tether PKA to defined cellular compartments establishing distinct pools to increase the specificity of PKA signalling. Here, we elucidated the structure of an extended PKA-binding domain of AKAP18beta bound to the D/D domain of the regulatory RIIalpha subunits of PKA. We identified three hydrophilic anchor points in AKAP18beta outside the core PKA-binding domain, which mediate contacts with the D/D domain. Such anchor points are conserved within AKAPs that bind regulatory RII subunits of PKA. We derived a different set of anchor points in AKAPs binding regulatory RI subunits of PKA. <em>In vitro</em> and cell-based experiments confirm the relevance of these sites for the interaction of RII subunits with AKAP18 and of RI subunits with the RI-specific smAKAP. Thus we report a novel mechanism governing interactions of AKAPs with PKA. The sequence specificity of each AKAP around the anchor points and the requirement of these points for the tight binding of PKA allow the development of selective inhibitors to unequivocally ascribe cellular functions to the AKAP18-PKA and other AKAP-PKA interactions. | |||
AKAP18:PKA-RIIalpha structure reveals crucial anchor points for recognition of regulatory subunits of PKA.,Gotz F, Roske Y, Schulz MS, Autenrieth K, Bertinetti D, Faelber K, Zuhlke K, Kreuchwig A, Kennedy E, Krause G, Daumke O, Herberg FW, Heinemann U, Klussmann E Biochem J. 2016 Apr 21. pii: BCJ20160242. PMID:27102985<ref>PMID:27102985</ref> | |||
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | |||
[[Category: | </div> | ||
[[Category: | <div class="pdbe-citations 4zp3" style="background-color:#fffaf0;"></div> | ||
[[Category: | |||
[[Category: | ==See Also== | ||
[[Category: | *[[A-kinase anchor protein|A-kinase anchor protein]] | ||
[[Category: | *[[A-kinase anchor protein 3D structures|A-kinase anchor protein 3D structures]] | ||
[[Category: Heinemann | *[[CAMP-dependent protein kinase 3D structures|CAMP-dependent protein kinase 3D structures]] | ||
[[Category: | == References == | ||
[[Category: | <references/> | ||
[[Category: | __TOC__ | ||
[[Category: | </StructureSection> | ||
[[Category: | [[Category: Homo sapiens]] | ||
[[Category: Large Structures]] | |||
[[Category: Autenrieth K]] | |||
[[Category: Daumke O]] | |||
[[Category: Faelber K]] | |||
[[Category: Goetz F]] | |||
[[Category: Heinemann U]] | |||
[[Category: Herberg FW]] | |||
[[Category: Klussmann E]] | |||
[[Category: Krause G]] | |||
[[Category: Kreuchwig A]] | |||
[[Category: Roske Y]] | |||
[[Category: Zuehlke K]] | |||
Latest revision as of 14:33, 9 May 2024
AKAP18:PKA-RIIalpha structure reveals crucial anchor points for recognition of regulatory subunits of PKAAKAP18:PKA-RIIalpha structure reveals crucial anchor points for recognition of regulatory subunits of PKA
Structural highlights
FunctionAKA7G_HUMAN Probably targets cAMP-dependent protein kinase (PKA) to the cellular membrane or cytoskeletal structures. The membrane-associated form reduces epithelial sodium channel (ENaC) activity, whereas the free cytoplasmic form may negatively regulate ENaC channel feedback inhibition by intracellular sodium.[1] [2] Publication Abstract from PubMedA-kinase anchoring proteins (AKAPs) interact with the dimerization/docking (D/D) domains of regulatory subunits of the ubiquitous protein kinase A (PKA). AKAPs tether PKA to defined cellular compartments establishing distinct pools to increase the specificity of PKA signalling. Here, we elucidated the structure of an extended PKA-binding domain of AKAP18beta bound to the D/D domain of the regulatory RIIalpha subunits of PKA. We identified three hydrophilic anchor points in AKAP18beta outside the core PKA-binding domain, which mediate contacts with the D/D domain. Such anchor points are conserved within AKAPs that bind regulatory RII subunits of PKA. We derived a different set of anchor points in AKAPs binding regulatory RI subunits of PKA. <em>In vitro</em> and cell-based experiments confirm the relevance of these sites for the interaction of RII subunits with AKAP18 and of RI subunits with the RI-specific smAKAP. Thus we report a novel mechanism governing interactions of AKAPs with PKA. The sequence specificity of each AKAP around the anchor points and the requirement of these points for the tight binding of PKA allow the development of selective inhibitors to unequivocally ascribe cellular functions to the AKAP18-PKA and other AKAP-PKA interactions. AKAP18:PKA-RIIalpha structure reveals crucial anchor points for recognition of regulatory subunits of PKA.,Gotz F, Roske Y, Schulz MS, Autenrieth K, Bertinetti D, Faelber K, Zuhlke K, Kreuchwig A, Kennedy E, Krause G, Daumke O, Herberg FW, Heinemann U, Klussmann E Biochem J. 2016 Apr 21. pii: BCJ20160242. PMID:27102985[3] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See Also
References
|
| ||||||||||||||||||