2vgb: Difference between revisions
No edit summary |
No edit summary |
||
| Line 22: | Line 22: | ||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=2vgb ConSurf]. | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=2vgb ConSurf]. | ||
<div style="clear:both"></div> | <div style="clear:both"></div> | ||
==See Also== | ==See Also== | ||
Latest revision as of 10:07, 1 May 2024
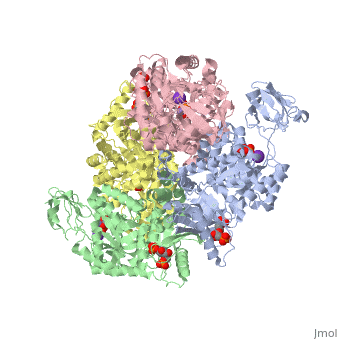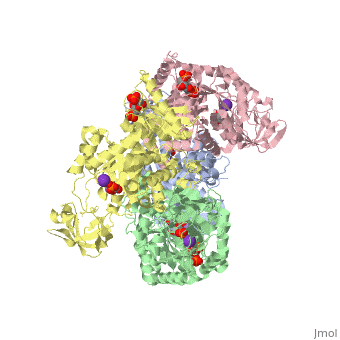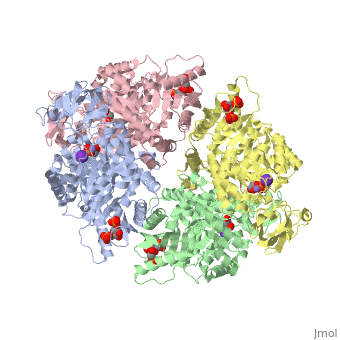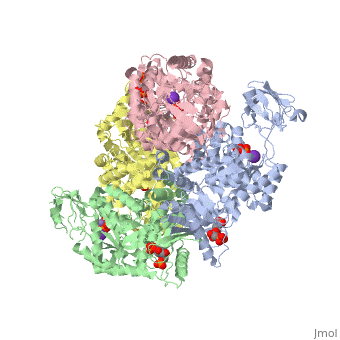
HUMAN ERYTHROCYTE PYRUVATE KINASEHUMAN ERYTHROCYTE PYRUVATE KINASE
Structural highlights
DiseaseKPYR_HUMAN Defects in PKLR are the cause of pyruvate kinase hyperactivity (PKHYP) [MIM:102900; also known as high red cell ATP syndrome. This autosomal dominant phenotype is characterized by increase of red blood cell ATP.[1] Defects in PKLR are the cause of pyruvate kinase deficiency of red cells (PKRD) [MIM:266200. A frequent cause of hereditary non-spherocytic hemolytic anemia. Clinically, pyruvate kinase-deficient patients suffer from a highly variable degree of chronic hemolysis, ranging from severe neonatal jaundice and fatal anemia at birth, severe transfusion-dependent chronic hemolysis, moderate hemolysis with exacerbation during infection, to a fully compensated hemolysis without apparent anemia. FunctionKPYR_HUMAN Plays a key role in glycolysis (By similarity). Evolutionary Conservation Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. See AlsoReferences
|
| ||||||||||||||||||