Amino Acids: Difference between revisions
Eric Martz (talk | contribs) |
Eric Martz (talk | contribs) |
||
| (29 intermediate revisions by 2 users not shown) | |||
| Line 3: | Line 3: | ||
==22 Standard Amino Acids and Mnemonics== | ==22 Standard Amino Acids and Mnemonics== | ||
Here are the names of the twenty standard amino acids, with their three and one-letter<ref>A one-letter notation for amino acid sequences (definitive rules). IUPAC—IUB Commission on Biochemical Nomenclature, pages 639-645, International Union of Pure and Applied Chemistry and International Union of Biochemistry, Butterworths, London, 1971. [ | Here are the names of the twenty-two standard amino acids, with their three and one-letter<ref>A one-letter notation for amino acid sequences (definitive rules). IUPAC—IUB Commission on Biochemical Nomenclature, pages 639-645, International Union of Pure and Applied Chemistry and International Union of Biochemistry, Butterworths, London, 1971. [https://old.iupac.org/publications/pac/1972/pdf/3104x0639.pdf Full Text].</ref> abbreviations. Mnemonic names are intended to help you to remember the one-letter codes, but are not the correct names. | ||
<center> | <center> | ||
| Line 20: | Line 20: | ||
</big></tt></td><td><tt><big> | </big></tt></td><td><tt><big> | ||
Lys K Lysine<br> (mnemonic: li<b>K</b>esine) | Lys K Lysine<br> (mnemonic: li<b>K</b>esine) | ||
<br> | <br> | ||
| Line 29: | Line 27: | ||
<br> | <br> | ||
Pro P <b>P</b>roline | Pro P <b>P</b>roline | ||
<br> | |||
Pyl O Pyrr<b>o</b>lysine* | |||
</big></tt></td><td rowspan="2"><tt><big> | </big></tt></td><td rowspan="2"><tt><big> | ||
| Line 79: | Line 79: | ||
<br> | <br> | ||
Ile I <b>I</b>soleucine | Ile I <b>I</b>soleucine | ||
<br> | |||
Leu L <b>L</b>eucine | |||
</big></tt></td><td><tt><big> | </big></tt></td><td><tt><big> | ||
Sec U Selenocysteine*<br> (mnemonic: seleni<b>U</b>m) | |||
<br> | |||
Ser S <b>S</b>erine | Ser S <b>S</b>erine | ||
<br> | <br> | ||
| Line 93: | Line 97: | ||
</tr> | </tr> | ||
<tr><td colspan="4"><center> | <tr><td colspan="4"><center> | ||
<nowiki>*</nowiki> | <nowiki>*</nowiki>Since the mid-20th century, there were 20 standard amino acids. Early in the 21st Century, | ||
[[#Unusual_Amino_Acids| | genetically encoded | ||
[[#Unusual_Amino_Acids|Selenocysteine and Pyrrolysine]] have been included as standard by the World Wide Protein Data Bank<ref name="pdb22" />. | |||
<br> | <br> | ||
† Asx: Asn or Asp; Glx: Gln or Glu; Unk: unknown. | † Asx: Asn or Asp; Glx: Gln or Glu; Unk: unknown. | ||
<br> | <br> | ||
[http:// | [http://old.iupac.org/publications/pac/1972/pdf/3104x0639.pdf IUPAC/IUB 1971 publication] defining one-letter codes. | ||
</center></td></tr> | </center></td></tr> | ||
</table> | </table> | ||
Click on the image below to see it full size | Click on the image below to see it full size with details on 21 amino acids. | ||
{| align="center" | {| align="center" | ||
| Line 119: | Line 123: | ||
==L- and D-Amino Acids== | ==L- and D-Amino Acids== | ||
<table border='1' cellpadding='3' align='right' hspace="8"><tr><td colspan='2'> | <table border='1' cellpadding='3' align='right' hspace="8" class="wikitable" style="margin:0px 0px 0px 8px;"><tr><td colspan='2'> | ||
PDB Nomenclature | PDB Nomenclature | ||
</td></tr><tr><td> | </td></tr><tr><td> | ||
| Line 127: | Line 131: | ||
</td></tr><tr><td> | </td></tr><tr><td> | ||
ALA<br>ARG<br>ASN<br>ASP<br>CYS<br>GLN<br>GLU<br>HIS<br>ILE<br>LEU<br>LYS<br>MET<br> | ALA<br>ARG<br>ASN<br>ASP<br>CYS<br>GLN<br>GLU<br>HIS<br>ILE<br>LEU<br>LYS<br>MET<br> | ||
PHE<br>PRO<br>SER<br>THR<br>TRP<br>TYR<br>VAL | PHE<br>PRO<br>PYL<br>SEC<br>SER<br>THR<br>TRP<br>TYR<br>VAL | ||
</td><td> | </td><td> | ||
DAL<br>DAR<br>DSG<br>DAS<br>DCY<br>DGN<br>DGL<br>DHI<br>DIL<br>DLE<br>DLY<br>MED<br> | DAL<br>DAR<br>DSG<br>DAS<br>DCY<br>DGN<br>DGL<br>DHI<br>DIL<br>DLE<br>DLY<br>MED<br> | ||
DPN<br>DPR<br>DSN<br>DTH<br>DTR<br>DTY<br>DVA | DPN<br>DPR<br>?<ref name="ddd">In January, 2022, no D-selenocysteine nor D-pyrrolysine were present in the [[wwPDB]], according to David Armstrong of PDBe.</ref><br>?<ref name="ddd" /><br>DSN<br>DTH<br>DTR<br>DTY<br>DVA | ||
</td></tr></table> | </td></tr></table> | ||
The asymmetric α-carbon in | The asymmetric α-carbon in 21 of the standard amino acids (all except glycine) is a chiral (stereogenic) center<ref name='chirality' />. Thus, each amino acid can exist in either of two optical isomers or enantiomers (mirror images), designated the L-form and D-form. Nearly all naturally-occuring amino acids are L-amino acids, so named because they are analogous to the L-form of glyceraldehyde<ref name='chirality'>See [http://en.wikipedia.org/wiki/Chirality_(chemistry) Chirality in Wikipedia]].</ref>. (Not all L-amino acids are levorotatory; some rotate polarized light clockwise<ref name='chirality' />). | ||
More than 700 entries in the [[PDB]] contain one or more D amino acids. Although D-amino acids are rare in nature, they do occur (see [http://en.wikipedia.org/wiki/Chirality_(chemistry)#D-Amino_Acid_Natural_Abundance natural abundance in Wikipedia]). D-alanine (DAL) occurs in over 100 entries in the [[PDB]], D-leucine (DLE), D-phenylalanine (DPN), D-serine (DSN), and D-valine (DVA) each occur in over 50. Some of these entries are chemically synthesized, not naturally occuring. Gramicidin is a naturally occuring antibiotic containing several D-amino acids, e.g. [[1nrm]]. [[4glu]] is the 102 amino acid chain of VEGF-A synthesized entirely from D-amino acids. | More than 700 entries in the [[PDB]] contain one or more D amino acids. Although D-amino acids are rare in nature, they do occur (see [http://en.wikipedia.org/wiki/Chirality_(chemistry)#D-Amino_Acid_Natural_Abundance natural abundance in Wikipedia]). D-alanine (DAL) occurs in over 100 entries in the [[PDB]], D-leucine (DLE), D-phenylalanine (DPN), D-serine (DSN), and D-valine (DVA) each occur in over 50. Some of these entries are chemically synthesized, not naturally occuring. Gramicidin is a naturally occuring antibiotic containing several D-amino acids, e.g. [[1nrm]]. [[4glu]] is the 102 amino acid chain of VEGF-A synthesized entirely from D-amino acids. | ||
| Line 139: | Line 143: | ||
==Unusual Amino Acids== | ==Unusual Amino Acids== | ||
There are at least two non-standard amino acids | There are at least two historically "non-standard" amino acids now recognized to be [http://en.wikipedia.org/wiki/Genetic_code genetically encoded], discussed below. In 2014, the [[PDB]] began including these two to make 22 standard amino acids<ref name="pdb22">[https://www.wwpdb.org/news/news?year=2014#5764490799cccf749a90cddf Announcement: Standardization of Amino Acid Nomenclature], World Wide Protein Data Bank News, January 8, 2014.</ref>. | ||
===Selenocysteine: the 21st amino acid=== | ===Selenocysteine: the 21st amino acid=== | ||
Rare proteins in all domains of life include '''[[Selenocysteine|selenocysteine (Sec, U)]]''', designated the 21st amino acid<ref name='21st'>PMID: 11028985</ref>. | Rare proteins in all domains of life include '''[[Selenocysteine|selenocysteine (Sec, U)]]''', designated the 21st amino acid<ref name='21st'>PMID: 11028985</ref>. In 2024, over 90 entries in the [[PDB]] include selenocysteine, identified as SEC. | ||
===Pyrrolysine: the 22nd amino acid=== | ===Pyrrolysine: the 22nd amino acid=== | ||
'''[[Pyrrolysine|pyrrolysine (Pyl, O)]]'''<ref>See [http://en.wikipedia.org/wiki/Pyrrolysine Pyrrolysine in Wikipedia].</ref> is a genetically encoded amino acid that occurs naturally in methanogenic archaea, | '''[[Pyrrolysine|pyrrolysine (Pyl, O)]]'''<ref>See [http://en.wikipedia.org/wiki/Pyrrolysine Pyrrolysine in Wikipedia].</ref> is a genetically encoded amino acid that occurs naturally in methanogenic archaea, now designated the 22nd amino acid<ref>PMID: 15380192</ref>. It occurs in [http://www.rcsb.org/pdb/ligand/ligandsummary.do?hetId=PYL several entries] in the PDB. | ||
See [[Pyrrolysine|more details]]. | See [[Pyrrolysine|more details]]. | ||
<!-- | <!-- | ||
| Line 162: | Line 166: | ||
===Postranslational modifications=== | ===Postranslational modifications=== | ||
Many unusual amino acids are formed by enzyme-catalyzed reactions that modify a standard amino acid after it has been included in a polypeptide; these are called post-translational modifications. Among them: | Many unusual amino acids are formed by enzyme-catalyzed reactions that modify a genetically encoded standard amino acid after it has been included in a polypeptide; these are called post-translational modifications. Among them: | ||
* phosphorylation of serine, threonine, or tyrosine | * phosphorylation of serine, threonine, or tyrosine | ||
** Example: C kinase [[1bkx]] | ** Example: C kinase [[1bkx]] | ||
| Line 178: | Line 182: | ||
==Structure, Properties, Behaviors in Proteins== | ==Structure, Properties, Behaviors in Proteins== | ||
Angel Herráez has provided a [http://biomodel.uah.es/en/model1/prot/aa-intro.htm tutorial introduction to amino acid structure] in which | Angel Herráez has provided a [http://biomodel.uah.es/en/model1/prot/aa-intro.htm tutorial introduction to amino acid structure] in which 20 amino acids may be visualized in 3D using JSmol. | ||
Please help to expand the list below to include all | Please help to expand the list below to include all 22 amino acids. | ||
===Glycine=== | ===Glycine=== | ||
| Line 218: | Line 222: | ||
*[[Standard Residues]] | *[[Standard Residues]] | ||
*[http://en.wikipedia.org/wiki/Amino_acids Amino Acids in Wikipedia] | *[http://en.wikipedia.org/wiki/Amino_acids Amino Acids in Wikipedia] | ||
*[ | *[[Basics of Protein Structure]] | ||
==References== | ==References== | ||
<references /> | <references /> | ||
Latest revision as of 23:34, 29 April 2024
For a general introduction to amino acids, please see Amino Acids in Wikipedia.
22 Standard Amino Acids and Mnemonics22 Standard Amino Acids and Mnemonics
Here are the names of the twenty-two standard amino acids, with their three and one-letter[1] abbreviations. Mnemonic names are intended to help you to remember the one-letter codes, but are not the correct names.
|
Ala A Alanine
|
Lys K Lysine |
mnemonic:
|
mnemonic:
|
|
Gln Q Glutamine |
Sec U Selenocysteine* |
||
*Since the mid-20th century, there were 20 standard amino acids. Early in the 21st Century,
genetically encoded
Selenocysteine and Pyrrolysine have been included as standard by the World Wide Protein Data Bank[2].
| |||
Click on the image below to see it full size with details on 21 amino acids.
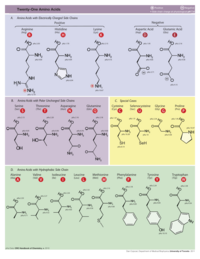 |
There is also a list of standard amino acids at the Protein Data Bank.
L- and D-Amino AcidsL- and D-Amino Acids
|
PDB Nomenclature | |
|
L-form |
D-form |
|
ALA |
DAL |
The asymmetric α-carbon in 21 of the standard amino acids (all except glycine) is a chiral (stereogenic) center[4]. Thus, each amino acid can exist in either of two optical isomers or enantiomers (mirror images), designated the L-form and D-form. Nearly all naturally-occuring amino acids are L-amino acids, so named because they are analogous to the L-form of glyceraldehyde[4]. (Not all L-amino acids are levorotatory; some rotate polarized light clockwise[4]).
More than 700 entries in the PDB contain one or more D amino acids. Although D-amino acids are rare in nature, they do occur (see natural abundance in Wikipedia). D-alanine (DAL) occurs in over 100 entries in the PDB, D-leucine (DLE), D-phenylalanine (DPN), D-serine (DSN), and D-valine (DVA) each occur in over 50. Some of these entries are chemically synthesized, not naturally occuring. Gramicidin is a naturally occuring antibiotic containing several D-amino acids, e.g. 1nrm. 4glu is the 102 amino acid chain of VEGF-A synthesized entirely from D-amino acids.
Unusual Amino AcidsUnusual Amino Acids
There are at least two historically "non-standard" amino acids now recognized to be genetically encoded, discussed below. In 2014, the PDB began including these two to make 22 standard amino acids[2].
Selenocysteine: the 21st amino acidSelenocysteine: the 21st amino acid
Rare proteins in all domains of life include selenocysteine (Sec, U), designated the 21st amino acid[5]. In 2024, over 90 entries in the PDB include selenocysteine, identified as SEC.
Pyrrolysine: the 22nd amino acidPyrrolysine: the 22nd amino acid
pyrrolysine (Pyl, O)[6] is a genetically encoded amino acid that occurs naturally in methanogenic archaea, now designated the 22nd amino acid[7]. It occurs in several entries in the PDB. See more details.
No 23rd amino acid?No 23rd amino acid?
The 21st and 22nd amino acids are specified by the stop codons UGA and UAG respectively, modified with downstream stem-loop structures in the mRNA. Lobanov et al.[8] searched "16 archaeal and 130 bacterial genomes for tRNAs with anticodons corresponding to the three stop signals". Their data suggest that "the occurrence of additional amino acids that are widely distributed and genetically encoded is unlikely."
SelenomethionineSelenomethionine
Please see Selenomethionine.
Postranslational modificationsPostranslational modifications
Many unusual amino acids are formed by enzyme-catalyzed reactions that modify a genetically encoded standard amino acid after it has been included in a polypeptide; these are called post-translational modifications. Among them:
- phosphorylation of serine, threonine, or tyrosine
- Example: C kinase 1bkx
- hydroxylation of proline (to yield hydroxyproline, e.g. in collagen)
- acetylation of lysine or the N terminus
- Example of N-terminal acetylation: 4mdh
- methylation of lysine
- carboxylation of aspartate or glutamate
- nitrosylation, e.g. to produce nitrotyrosine
- acylation with a fatty acid (see N-terminal myristoylation of Recoverin)
- prenylation
- glycosylation of serine, threonine or asparagine (yielding glycoproteins)
- Example of N-linked glycolylation: 1igy
Structure, Properties, Behaviors in ProteinsStructure, Properties, Behaviors in Proteins
Angel Herráez has provided a tutorial introduction to amino acid structure in which 20 amino acids may be visualized in 3D using JSmol.
Please help to expand the list below to include all 22 amino acids.
GlycineGlycine
Gly G: Small.
See also Glycine in Proteopedia and Glycine in Wikipedia, where the structure is shown. Glycine is the smallest amino acid, since its side-chain is nothing but a single hydrogen. Glycine is frequently found in turns, since the steric clashes of larger amino acids would be problematic. Turns typically occur on the surfaces of proteins, between beta-strands or alpha-helices. It is common to find highly conserved glycines in turns on protein surfaces, since mutation to a bulkier residue would interfere with the fold. Glycine also occurs in beta-strands and alpha-helices, although its frequency in alpha helices is low (second lowest, after proline, p. 125 in [9]). Glycine is commonly found at the C-terminus of alpha helices, and is considered a helix terminator (p. 125 in [9]).
HistidineHistidine
His H: Charged: Basic, Aromatic, Bulky
See Histidine in Wikipedia, where the structure is shown. The sidechain of His can be positively charged (protonated), in which case both of the nitrogens in the sidechain imidazole ring have hydrogens, and the charge is delocalized between them. The pKa for protonation is 6.1. This means that, on average at any moment, half of the His sidechains are protonated when the pH is 6.1. At the pH of blood, 7.4 ±0.05, His sidechains are positively charged less than 10% of the time. Therefore, His is not included in the usual list of positively charged amino acids (lysine and arginine).
PhenylalaninePhenylalanine
Phe F: Neutral, Aromatic, Bulky
See Phenylalanine in Wikipedia, where the structure is shown. Phe participates in Cation-pi interactions. Phenylketonuria is a genetic disease in which the enzyme that converts phenylalanine to tyrosine is nonfunctional, leading to toxicity from excess phenylalanine, and tyrosine deficiency.
TryptophanTryptophan
Trp W: Neutral, Polar, Aromatic, Bulky
See Tryptophan in Wikipedia, where the structure is shown. In trans-membrane proteins, tryptophans often lie at the interface between water and lipid. An example is the potassium channel, e.g. 1bl8. To see their positions, use the Find dialog in FirstGlance in Jmol. Trp participates in Cation-pi interactions.
TyrosineTyrosine
Tyr Y: Neutral, Polar, Aromatic, Bulky
See Tyrosine in Wikipedia, where the structure is shown, and Nitrotyrosine. Tyr participates in Cation-pi interactions.
See AlsoSee Also
ReferencesReferences
- ↑ A one-letter notation for amino acid sequences (definitive rules). IUPAC—IUB Commission on Biochemical Nomenclature, pages 639-645, International Union of Pure and Applied Chemistry and International Union of Biochemistry, Butterworths, London, 1971. Full Text.
- ↑ 2.0 2.1 Announcement: Standardization of Amino Acid Nomenclature, World Wide Protein Data Bank News, January 8, 2014.
- ↑ 3.0 3.1 In January, 2022, no D-selenocysteine nor D-pyrrolysine were present in the wwPDB, according to David Armstrong of PDBe.
- ↑ 4.0 4.1 4.2 See Chirality in Wikipedia].
- ↑ Atkins JF, Gesteland RF. The twenty-first amino acid. Nature. 2000 Sep 28;407(6803):463, 465. PMID:11028985 doi:10.1038/35035189
- ↑ See Pyrrolysine in Wikipedia.
- ↑ Hao B, Zhao G, Kang PT, Soares JA, Ferguson TK, Gallucci J, Krzycki JA, Chan MK. Reactivity and chemical synthesis of L-pyrrolysine- the 22(nd) genetically encoded amino acid. Chem Biol. 2004 Sep;11(9):1317-24. PMID:15380192 doi:10.1016/j.chembiol.2004.07.011
- ↑ Lobanov AV, Kryukov GV, Hatfield DL, Gladyshev VN. Is there a twenty third amino acid in the genetic code? Trends Genet. 2006 Jul;22(7):357-60. Epub 2006 May 19. PMID:16713651 doi:10.1016/j.tig.2006.05.002
- ↑ 9.0 9.1 Kessel, Amit, and Nir Ben-Tal. Introduction to Proteins: Structure, Function, and Motion. CRC Press, 2011. 653 pages.