1gos: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
No edit summary |
||
| (2 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
==Human Monoamine Oxidase B== | ==Human Monoamine Oxidase B== | ||
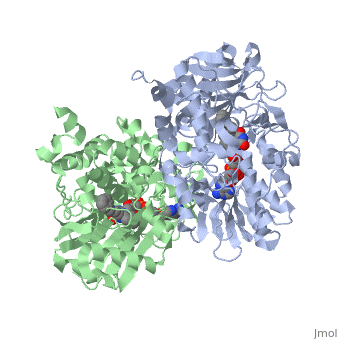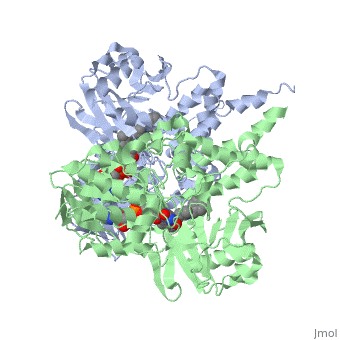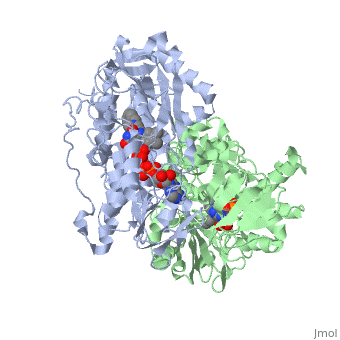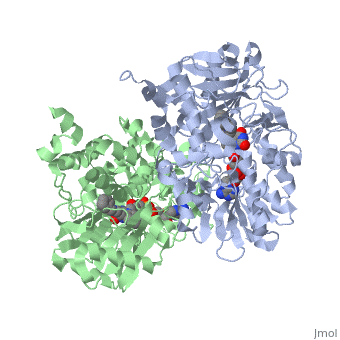
<StructureSection load='1gos' size='340' side='right' caption='[[1gos]], [[Resolution|resolution]] 3.00Å' scene=''> | <StructureSection load='1gos' size='340' side='right'caption='[[1gos]], [[Resolution|resolution]] 3.00Å' scene=''> | ||
== Structural highlights == | == Structural highlights == | ||
<table><tr><td colspan='2'>[[1gos]] is a 2 chain structure with sequence from [ | <table><tr><td colspan='2'>[[1gos]] is a 2 chain structure with sequence from [https://en.wikipedia.org/wiki/Homo_sapiens Homo sapiens]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1GOS OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=1GOS FirstGlance]. <br> | ||
</td></tr><tr id=' | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 3Å</td></tr> | ||
<tr id=' | <tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=FAD:FLAVIN-ADENINE+DINUCLEOTIDE'>FAD</scene>, <scene name='pdbligand=NYP:N-[(E)-METHYL](PHENYL)-N-[(E)-2-PROPENYLIDENE]METHANAMINIUM'>NYP</scene></td></tr> | ||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1gos FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1gos OCA], [https://pdbe.org/1gos PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1gos RCSB], [https://www.ebi.ac.uk/pdbsum/1gos PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1gos ProSAT]</span></td></tr> | |||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[ | |||
</table> | </table> | ||
== Function == | |||
[https://www.uniprot.org/uniprot/AOFB_HUMAN AOFB_HUMAN] Catalyzes the oxidative deamination of biogenic and xenobiotic amines and has important functions in the metabolism of neuroactive and vasoactive amines in the central nervous system and peripheral tissues. MAOB preferentially degrades benzylamine and phenylethylamine. | |||
== Evolutionary Conservation == | == Evolutionary Conservation == | ||
[[Image:Consurf_key_small.gif|200px|right]] | [[Image:Consurf_key_small.gif|200px|right]] | ||
| Line 19: | Line 20: | ||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1gos ConSurf]. | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1gos ConSurf]. | ||
<div style="clear:both"></div> | <div style="clear:both"></div> | ||
==See Also== | ==See Also== | ||
*[[Monoamine oxidase | *[[Monoamine oxidase|Monoamine oxidase]] | ||
__TOC__ | __TOC__ | ||
</StructureSection> | </StructureSection> | ||
[[Category: | [[Category: Homo sapiens]] | ||
[[Category: | [[Category: Large Structures]] | ||
[[Category: Binda | [[Category: Binda C]] | ||
[[Category: Edmondson | [[Category: Edmondson DE]] | ||
[[Category: Hubalek | [[Category: Hubalek F]] | ||
[[Category: Mattevi | [[Category: Mattevi A]] | ||
[[Category: Newton-Vinson | [[Category: Newton-Vinson P]] | ||
Latest revision as of 14:23, 27 March 2024
Human Monoamine Oxidase BHuman Monoamine Oxidase B
Structural highlights
FunctionAOFB_HUMAN Catalyzes the oxidative deamination of biogenic and xenobiotic amines and has important functions in the metabolism of neuroactive and vasoactive amines in the central nervous system and peripheral tissues. MAOB preferentially degrades benzylamine and phenylethylamine. Evolutionary Conservation Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. See Also |
| ||||||||||||||||||