3rmv: Difference between revisions
Jump to navigation
Jump to search
New page: '''Unreleased structure''' The entry 3rmv is ON HOLD Authors: Chaikuad, A., Froese, D.S., Yue, W. W., Krysztofinska, E., von Delft, F., Weigelt, J., Arrowsmith, C.H., Edwards, A.M., Bou... |
No edit summary |
||
| (10 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
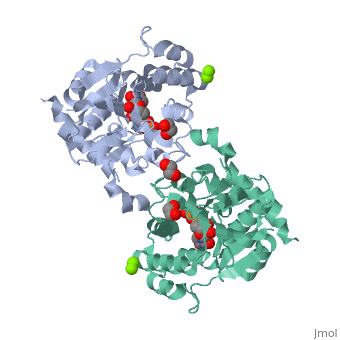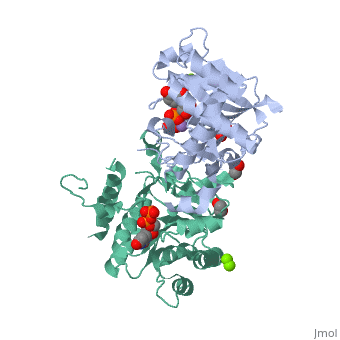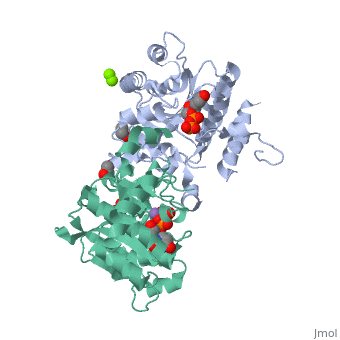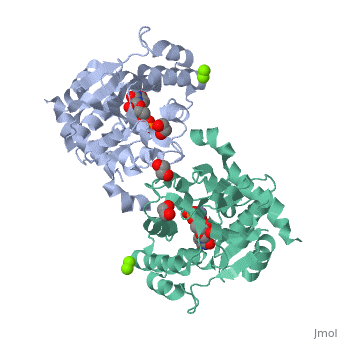
The entry | ==Crystal Structure of Human Glycogenin-1 (GYG1) T83M mutant complexed with manganese and UDP== | ||
<StructureSection load='3rmv' size='340' side='right'caption='[[3rmv]], [[Resolution|resolution]] 1.82Å' scene=''> | |||
== Structural highlights == | |||
<table><tr><td colspan='2'>[[3rmv]] is a 1 chain structure with sequence from [https://en.wikipedia.org/wiki/Homo_sapiens Homo sapiens]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=3RMV OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=3RMV FirstGlance]. <br> | |||
</td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 1.82Å</td></tr> | |||
<tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=EDO:1,2-ETHANEDIOL'>EDO</scene>, <scene name='pdbligand=MG:MAGNESIUM+ION'>MG</scene>, <scene name='pdbligand=MN:MANGANESE+(II)+ION'>MN</scene>, <scene name='pdbligand=UDP:URIDINE-5-DIPHOSPHATE'>UDP</scene></td></tr> | |||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=3rmv FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=3rmv OCA], [https://pdbe.org/3rmv PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=3rmv RCSB], [https://www.ebi.ac.uk/pdbsum/3rmv PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=3rmv ProSAT]</span></td></tr> | |||
</table> | |||
== Disease == | |||
[https://www.uniprot.org/uniprot/GLYG_HUMAN GLYG_HUMAN] Glycogen storage disease due to glycogenin deficiency. The disease is caused by mutations affecting the gene represented in this entry. The disease is caused by mutations affecting the gene represented in this entry. | |||
== Function == | |||
[https://www.uniprot.org/uniprot/GLYG_HUMAN GLYG_HUMAN] Self-glucosylates, via an inter-subunit mechanism, to form an oligosaccharide primer that serves as substrate for glycogen synthase. | |||
==See Also== | |||
*[[Glycogenin|Glycogenin]] | |||
__TOC__ | |||
</StructureSection> | |||
[[Category: Homo sapiens]] | |||
[[Category: Large Structures]] | |||
[[Category: Arrowsmith CH]] | |||
[[Category: Bountra C]] | |||
[[Category: Chaikuad A]] | |||
[[Category: Edwards AM]] | |||
[[Category: Froese DS]] | |||
[[Category: Krysztofinska E]] | |||
[[Category: Oppermann U]] | |||
[[Category: Weigelt J]] | |||
[[Category: Yue WW]] | |||
[[Category: Von Delft F]] | |||
Latest revision as of 15:29, 14 March 2024
Crystal Structure of Human Glycogenin-1 (GYG1) T83M mutant complexed with manganese and UDPCrystal Structure of Human Glycogenin-1 (GYG1) T83M mutant complexed with manganese and UDP
Structural highlights
DiseaseGLYG_HUMAN Glycogen storage disease due to glycogenin deficiency. The disease is caused by mutations affecting the gene represented in this entry. The disease is caused by mutations affecting the gene represented in this entry. FunctionGLYG_HUMAN Self-glucosylates, via an inter-subunit mechanism, to form an oligosaccharide primer that serves as substrate for glycogen synthase. See Also |
| ||||||||||||||||||