4q3h: Difference between revisions
No edit summary |
No edit summary |
||
| (One intermediate revision by the same user not shown) | |||
| Line 1: | Line 1: | ||
==The crystal structure of NHERF1 PDZ2 CXCR2 complex revealed by the NHERF1 CXCR2 chimeric protein== | ==The crystal structure of NHERF1 PDZ2 CXCR2 complex revealed by the NHERF1 CXCR2 chimeric protein== | ||
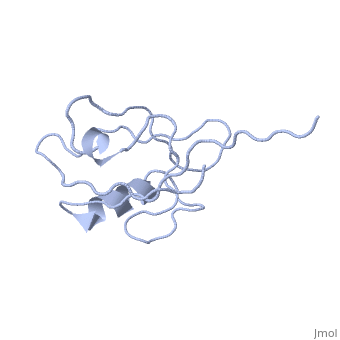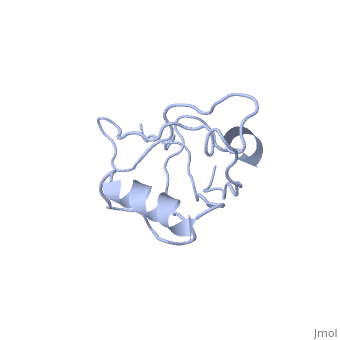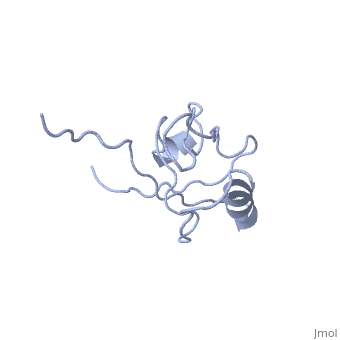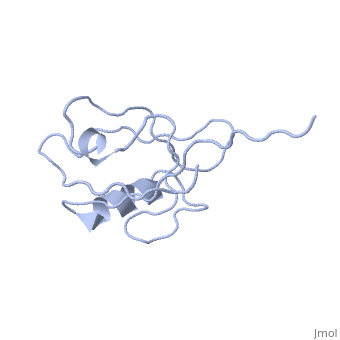
<StructureSection load='4q3h' size='340' side='right' caption='[[4q3h]], [[Resolution|resolution]] 1.44Å' scene=''> | <StructureSection load='4q3h' size='340' side='right'caption='[[4q3h]], [[Resolution|resolution]] 1.44Å' scene=''> | ||
== Structural highlights == | == Structural highlights == | ||
<table><tr><td colspan='2'>[[4q3h]] is a 2 chain structure with sequence from [ | <table><tr><td colspan='2'>[[4q3h]] is a 2 chain structure with sequence from [https://en.wikipedia.org/wiki/Homo_sapiens Homo sapiens]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=4Q3H OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=4Q3H FirstGlance]. <br> | ||
</td></tr><tr id=' | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 1.443Å</td></tr> | ||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[ | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=4q3h FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=4q3h OCA], [https://pdbe.org/4q3h PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=4q3h RCSB], [https://www.ebi.ac.uk/pdbsum/4q3h PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=4q3h ProSAT]</span></td></tr> | ||
</table> | </table> | ||
== Disease == | == Disease == | ||
[ | [https://www.uniprot.org/uniprot/NHRF1_HUMAN NHRF1_HUMAN] Defects in SLC9A3R1 are the cause of hypophosphatemic nephrolithiasis/osteoporosis type 2 (NPHLOP2) [MIM:[https://omim.org/entry/612287 612287]. Hypophosphatemia results from idiopathic renal phosphate loss. It contributes to the pathogenesis of hypophosphatemic urolithiasis (formation of urinary calculi) as well to that of hypophosphatemic osteoporosis (bone demineralization).<ref>PMID:18784102</ref> <ref>PMID:22506049</ref> | ||
== Function == | == Function == | ||
[ | [https://www.uniprot.org/uniprot/NHRF1_HUMAN NHRF1_HUMAN] Scaffold protein that connects plasma membrane proteins with members of the ezrin/moesin/radixin family and thereby helps to link them to the actin cytoskeleton and to regulate their surface expression. Necessary for recycling of internalized ADRB2. Was first known to play a role in the regulation of the activity and subcellular location of SLC9A3. Necessary for cAMP-mediated phosphorylation and inhibition of SLC9A3. May enhance Wnt signaling. May participate in HTR4 targeting to microvilli (By similarity). Involved in the regulation of phosphate reabsorption in the renal proximal tubules.<ref>PMID:9430655</ref> <ref>PMID:9096337</ref> <ref>PMID:10499588</ref> <ref>PMID:18784102</ref> | ||
==See Also== | |||
*[[Sodium-hydrogen exchange regulatory factor|Sodium-hydrogen exchange regulatory factor]] | |||
*[[Sodium-hydrogen exchange regulatory factor 3D structures|Sodium-hydrogen exchange regulatory factor 3D structures]] | |||
== References == | == References == | ||
<references/> | <references/> | ||
__TOC__ | __TOC__ | ||
</StructureSection> | </StructureSection> | ||
[[Category: | [[Category: Homo sapiens]] | ||
[[Category: Brunzelle | [[Category: Large Structures]] | ||
[[Category: Holcomb | [[Category: Brunzelle J]] | ||
[[Category: Jiang | [[Category: Holcomb J]] | ||
[[Category: Li | [[Category: Jiang Y]] | ||
[[Category: Lu | [[Category: Li C]] | ||
[[Category: Sirinupong | [[Category: Lu G]] | ||
[[Category: Trescott | [[Category: Sirinupong N]] | ||
[[Category: Yang | [[Category: Trescott L]] | ||
[[Category: Yang Z]] | |||
Latest revision as of 15:43, 1 March 2024
The crystal structure of NHERF1 PDZ2 CXCR2 complex revealed by the NHERF1 CXCR2 chimeric proteinThe crystal structure of NHERF1 PDZ2 CXCR2 complex revealed by the NHERF1 CXCR2 chimeric protein
Structural highlights
DiseaseNHRF1_HUMAN Defects in SLC9A3R1 are the cause of hypophosphatemic nephrolithiasis/osteoporosis type 2 (NPHLOP2) [MIM:612287. Hypophosphatemia results from idiopathic renal phosphate loss. It contributes to the pathogenesis of hypophosphatemic urolithiasis (formation of urinary calculi) as well to that of hypophosphatemic osteoporosis (bone demineralization).[1] [2] FunctionNHRF1_HUMAN Scaffold protein that connects plasma membrane proteins with members of the ezrin/moesin/radixin family and thereby helps to link them to the actin cytoskeleton and to regulate their surface expression. Necessary for recycling of internalized ADRB2. Was first known to play a role in the regulation of the activity and subcellular location of SLC9A3. Necessary for cAMP-mediated phosphorylation and inhibition of SLC9A3. May enhance Wnt signaling. May participate in HTR4 targeting to microvilli (By similarity). Involved in the regulation of phosphate reabsorption in the renal proximal tubules.[3] [4] [5] [6] See AlsoReferences
|
| ||||||||||||||||