1yv0: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
No edit summary |
||
| (6 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
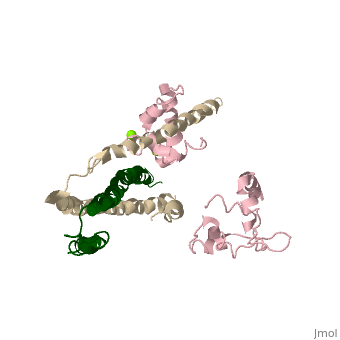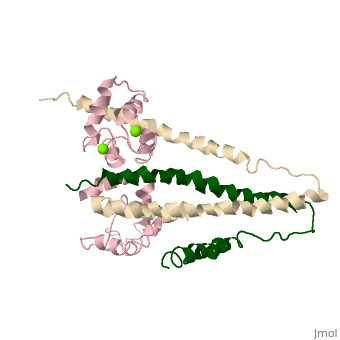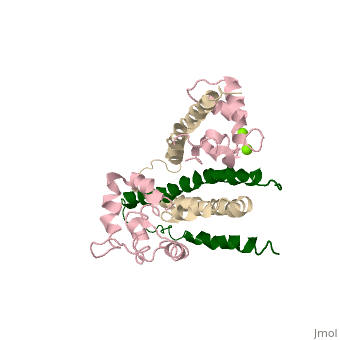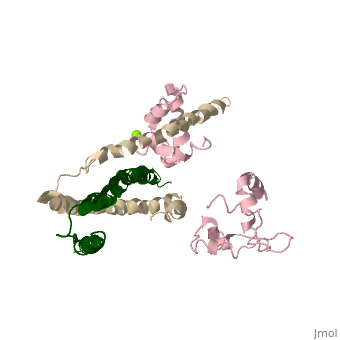
==Crystal structure of skeletal muscle troponin in the Ca2+-free state== | ==Crystal structure of skeletal muscle troponin in the Ca2+-free state== | ||
<StructureSection load='1yv0' size='340' side='right' caption='[[1yv0]], [[Resolution|resolution]] 7.00Å' scene=''> | <StructureSection load='1yv0' size='340' side='right'caption='[[1yv0]], [[Resolution|resolution]] 7.00Å' scene=''> | ||
== Structural highlights == | == Structural highlights == | ||
<table><tr><td colspan='2'>[[1yv0]] is a 3 chain structure with sequence from [ | <table><tr><td colspan='2'>[[1yv0]] is a 3 chain structure with sequence from [https://en.wikipedia.org/wiki/Gallus_gallus Gallus gallus]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1YV0 OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=1YV0 FirstGlance]. <br> | ||
</td></tr><tr><td class="sblockLbl"><b>[[ | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 7Å</td></tr> | ||
<tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=MG:MAGNESIUM+ION'>MG</scene></td></tr> | |||
<tr><td class="sblockLbl"><b>[[ | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1yv0 FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1yv0 OCA], [https://pdbe.org/1yv0 PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1yv0 RCSB], [https://www.ebi.ac.uk/pdbsum/1yv0 PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1yv0 ProSAT]</span></td></tr> | ||
<tr><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[ | </table> | ||
<table> | == Function == | ||
[https://www.uniprot.org/uniprot/TNNT3_CHICK TNNT3_CHICK] Troponin T is the tropomyosin-binding subunit of troponin, the thin filament regulatory complex which confers calcium-sensitivity to striated muscle actomyosin ATPase activity. | |||
== Evolutionary Conservation == | == Evolutionary Conservation == | ||
[[Image:Consurf_key_small.gif|200px|right]] | [[Image:Consurf_key_small.gif|200px|right]] | ||
Check<jmol> | Check<jmol> | ||
<jmolCheckbox> | <jmolCheckbox> | ||
<scriptWhenChecked>select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/yv/1yv0_consurf.spt"</scriptWhenChecked> | <scriptWhenChecked>; select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/yv/1yv0_consurf.spt"</scriptWhenChecked> | ||
<scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> | <scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> | ||
<text>to colour the structure by Evolutionary Conservation</text> | <text>to colour the structure by Evolutionary Conservation</text> | ||
</jmolCheckbox> | </jmolCheckbox> | ||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/ | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1yv0 ConSurf]. | ||
<div style="clear:both"></div> | <div style="clear:both"></div> | ||
==See Also== | ==See Also== | ||
*[[Troponin|Troponin]] | *[[Troponin|Troponin]] | ||
*[[Troponin 3D structures|Troponin 3D structures]] | |||
__TOC__ | __TOC__ | ||
</StructureSection> | </StructureSection> | ||
[[Category: Gallus gallus]] | [[Category: Gallus gallus]] | ||
[[Category: Cooke | [[Category: Large Structures]] | ||
[[Category: Fletterick | [[Category: Cooke R]] | ||
[[Category: Karatzaferi | [[Category: Fletterick RJ]] | ||
[[Category: Malanina | [[Category: Karatzaferi C]] | ||
[[Category: Mendelson | [[Category: Malanina GG]] | ||
[[Category: Stone | [[Category: Mendelson RA]] | ||
[[Category: Vinogradova | [[Category: Stone DB]] | ||
[[Category: Vinogradova MV]] | |||
Latest revision as of 12:00, 14 February 2024
Crystal structure of skeletal muscle troponin in the Ca2+-free stateCrystal structure of skeletal muscle troponin in the Ca2+-free state
Structural highlights
FunctionTNNT3_CHICK Troponin T is the tropomyosin-binding subunit of troponin, the thin filament regulatory complex which confers calcium-sensitivity to striated muscle actomyosin ATPase activity. Evolutionary Conservation Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. See Also |
| ||||||||||||||||||