4ct0: Difference between revisions
No edit summary |
No edit summary |
||
| Line 4: | Line 4: | ||
== Structural highlights == | == Structural highlights == | ||
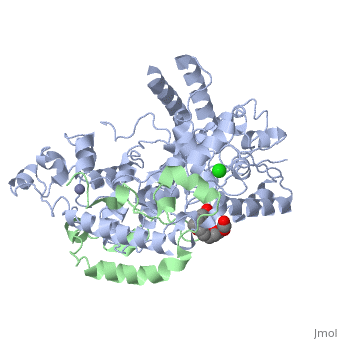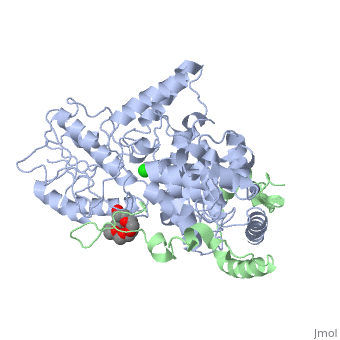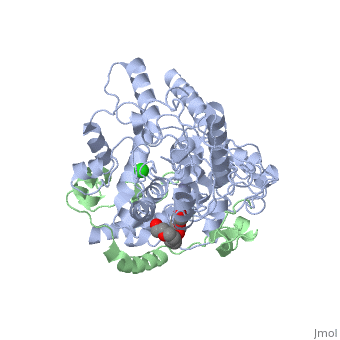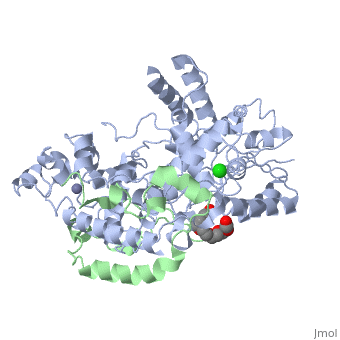
<table><tr><td colspan='2'>[[4ct0]] is a 2 chain structure with sequence from [https://en.wikipedia.org/wiki/Mus_musculus Mus musculus]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=4CT0 OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=4CT0 FirstGlance]. <br> | <table><tr><td colspan='2'>[[4ct0]] is a 2 chain structure with sequence from [https://en.wikipedia.org/wiki/Mus_musculus Mus musculus]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=4CT0 OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=4CT0 FirstGlance]. <br> | ||
</td></tr><tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=CL:CHLORIDE+ION'>CL</scene>, <scene name='pdbligand=P6G:HEXAETHYLENE+GLYCOL'>P6G</scene>, <scene name='pdbligand=ZN:ZINC+ION'>ZN</scene></td></tr> | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 2.45Å</td></tr> | ||
<tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=CL:CHLORIDE+ION'>CL</scene>, <scene name='pdbligand=P6G:HEXAETHYLENE+GLYCOL'>P6G</scene>, <scene name='pdbligand=ZN:ZINC+ION'>ZN</scene></td></tr> | |||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=4ct0 FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=4ct0 OCA], [https://pdbe.org/4ct0 PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=4ct0 RCSB], [https://www.ebi.ac.uk/pdbsum/4ct0 PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=4ct0 ProSAT]</span></td></tr> | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=4ct0 FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=4ct0 OCA], [https://pdbe.org/4ct0 PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=4ct0 RCSB], [https://www.ebi.ac.uk/pdbsum/4ct0 PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=4ct0 ProSAT]</span></td></tr> | ||
</table> | </table> | ||
== Function == | == Function == | ||
[https://www.uniprot.org/uniprot/CRY1_MOUSE CRY1_MOUSE] Blue light-dependent regulator of the circadian feedback loop. Inhibits CLOCK|NPAS2-ARNTL E box-mediated transcription. Acts, in conjunction with CRY2, in maintaining period length and circadian rhythmicity. Has no photolyase activity. Capable of translocating circadian clock core proteins such as PER proteins to the nucleus. May inhibit CLOCK|NPAS2-ARNTL transcriptional activity through stabilizing the unphosphorylated form of ARNTL.<ref>PMID:10428031</ref> <ref>PMID:16628007</ref> <ref>PMID:16478995</ref> | |||
<div style="background-color:#fffaf0;"> | <div style="background-color:#fffaf0;"> | ||
== Publication Abstract from PubMed == | == Publication Abstract from PubMed == | ||
Latest revision as of 15:16, 20 December 2023
Crystal Structure of Mouse Cryptochrome1 in Complex with Period2Crystal Structure of Mouse Cryptochrome1 in Complex with Period2
Structural highlights
FunctionCRY1_MOUSE Blue light-dependent regulator of the circadian feedback loop. Inhibits CLOCK|NPAS2-ARNTL E box-mediated transcription. Acts, in conjunction with CRY2, in maintaining period length and circadian rhythmicity. Has no photolyase activity. Capable of translocating circadian clock core proteins such as PER proteins to the nucleus. May inhibit CLOCK|NPAS2-ARNTL transcriptional activity through stabilizing the unphosphorylated form of ARNTL.[1] [2] [3] Publication Abstract from PubMedPeriod (PER) proteins are essential components of the mammalian circadian clock. They form complexes with cryptochromes (CRY), which negatively regulate CLOCK/BMAL1-dependent transactivation of clock and clock-controlled genes. To define the roles of mammalian CRY/PER complexes in the circadian clock, we have determined the crystal structure of a complex comprising the photolyase homology region of mouse CRY1 (mCRY1) and a C-terminal mouse PER2 (mPER2) fragment. mPER2 winds around the helical mCRY1 domain covering the binding sites of FBXL3 and CLOCK/BMAL1, but not the FAD binding pocket. Our structure revealed an unexpected zinc ion in one interface, which stabilizes mCRY1-mPER2 interactions in vivo. We provide evidence that mCRY1/mPER2 complex formation is modulated by an interplay of zinc binding and mCRY1 disulfide bond formation, which may be influenced by the redox state of the cell. Our studies may allow for the development of circadian and metabolic modulators. Interaction of Circadian Clock Proteins CRY1 and PER2 Is Modulated by Zinc Binding and Disulfide Bond Formation.,Schmalen I, Reischl S, Wallach T, Klemz R, Grudziecki A, Prabu JR, Benda C, Kramer A, Wolf E Cell. 2014 May 22;157(5):1203-15. doi: 10.1016/j.cell.2014.03.057. PMID:24855952[4] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences
|
| ||||||||||||||||||