1w68: Difference between revisions
No edit summary |
No edit summary |
||
| (13 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
==Crystal Structure of Mouse Ribonucleotide Reductase Subunit R2 under Oxidizing Conditions. A Fully Occupied Dinuclear Iron Cluster.== | |||
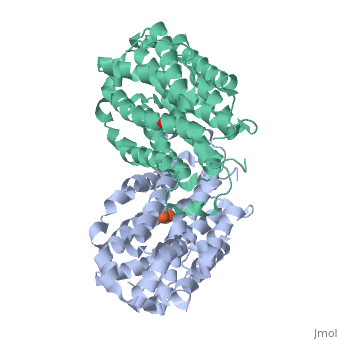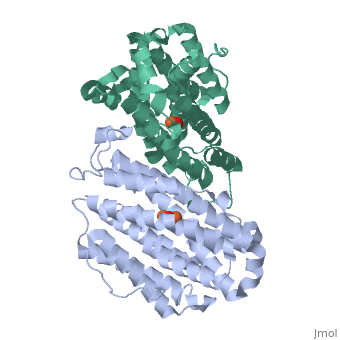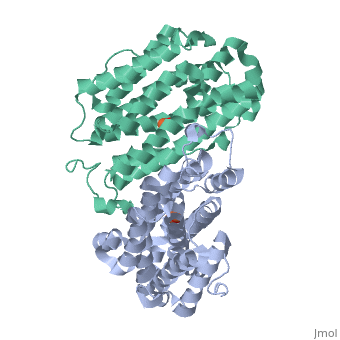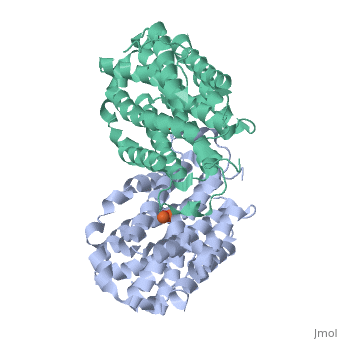
<StructureSection load='1w68' size='340' side='right'caption='[[1w68]], [[Resolution|resolution]] 2.20Å' scene=''> | |||
== Structural highlights == | |||
<table><tr><td colspan='2'>[[1w68]] is a 1 chain structure with sequence from [https://en.wikipedia.org/wiki/Mus_musculus Mus musculus]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1W68 OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=1W68 FirstGlance]. <br> | |||
</td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 2.2Å</td></tr> | |||
<tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=FEO:MU-OXO-DIIRON'>FEO</scene></td></tr> | |||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1w68 FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1w68 OCA], [https://pdbe.org/1w68 PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1w68 RCSB], [https://www.ebi.ac.uk/pdbsum/1w68 PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1w68 ProSAT]</span></td></tr> | |||
</table> | |||
== Function == | |||
[https://www.uniprot.org/uniprot/RIR2_MOUSE RIR2_MOUSE] Provides the precursors necessary for DNA synthesis. Catalyzes the biosynthesis of deoxyribonucleotides from the corresponding ribonucleotides. Inhibits Wnt signaling (By similarity). | |||
== Evolutionary Conservation == | |||
[[Image:Consurf_key_small.gif|200px|right]] | |||
Check<jmol> | |||
<jmolCheckbox> | |||
<scriptWhenChecked>; select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/w6/1w68_consurf.spt"</scriptWhenChecked> | |||
<scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> | |||
<text>to colour the structure by Evolutionary Conservation</text> | |||
</jmolCheckbox> | |||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1w68 ConSurf]. | |||
<div style="clear:both"></div> | |||
<div style="background-color:#fffaf0;"> | |||
== Publication Abstract from PubMed == | |||
Class I ribonucleotide reductase (RNR) catalyzes the de novo synthesis of deoxyribonucleotides in mammals and many other organisms. The RNR subunit R2 contains a dinuclear iron center, which in its diferrous form spontaneously reacts with O2, forming a mu-oxo-bridged diferric cluster and a stable tyrosyl radical. Here, we present the first crystal structures of R2 from mouse with its native dinuclear iron center, both under reducing and oxidizing conditions. In one structure obtained under reducing conditions, the iron-bridging ligand Glu-267 adopts the mu-(eta1,eta2) coordination mode, which has previously been related to O2 activation, and an acetate ion from the soaking solution is observed where O2 has been proposed to bind the iron. The structure of mouse R2 under oxidizing conditions resembles the nonradical diferric R2 from Escherichia coli, with the exception of the coordination of water and Asp-139 to Fe1. There are also additional water molecules near the tyrosyl radical site, as suggested by previous spectroscopic studies. Since no crystal structure of the active radical form has been reported, we propose models for the movement of waters and/or tyrosyl radical site when diferric R2 is oxidized to the radical form, in agreement with our previous ENDOR study. Compared with E. coli R2, two conserved phenylalanine residues in the hydrophobic environment around the diiron center have opposing rotameric conformations, and the carboxylate ligands of the diiron center in mouse R2 appear more flexible. Together, this might contribute to the lower affinity and cooperative binding of iron in mouse R2. | |||
Crystal structural studies of changes in the native dinuclear iron center of ribonucleotide reductase protein R2 from mouse.,Strand KR, Karlsen S, Kolberg M, Rohr AK, Gorbitz CH, Andersson KK J Biol Chem. 2004 Nov 5;279(45):46794-801. Epub 2004 Aug 17. PMID:15322079<ref>PMID:15322079</ref> | |||
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | |||
</div> | |||
<div class="pdbe-citations 1w68" style="background-color:#fffaf0;"></div> | |||
== | ==See Also== | ||
*[[Ribonucleotide reductase 3D structures|Ribonucleotide reductase 3D structures]] | |||
== References == | |||
<references/> | |||
__TOC__ | |||
</StructureSection> | |||
[[Category: Large Structures]] | |||
[[Category: Mus musculus]] | [[Category: Mus musculus]] | ||
[[Category: Andersson KK]] | |||
[[Category: Gorbitz CH]] | |||
[[Category: Andersson | [[Category: Karlsen S]] | ||
[[Category: Gorbitz | [[Category: Kolberg M]] | ||
[[Category: Karlsen | [[Category: Rohr AK]] | ||
[[Category: Kolberg | [[Category: Strand KR]] | ||
[[Category: Rohr | |||
[[Category: Strand | |||
Latest revision as of 16:17, 13 December 2023
Crystal Structure of Mouse Ribonucleotide Reductase Subunit R2 under Oxidizing Conditions. A Fully Occupied Dinuclear Iron Cluster.Crystal Structure of Mouse Ribonucleotide Reductase Subunit R2 under Oxidizing Conditions. A Fully Occupied Dinuclear Iron Cluster.
Structural highlights
FunctionRIR2_MOUSE Provides the precursors necessary for DNA synthesis. Catalyzes the biosynthesis of deoxyribonucleotides from the corresponding ribonucleotides. Inhibits Wnt signaling (By similarity). Evolutionary Conservation Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. Publication Abstract from PubMedClass I ribonucleotide reductase (RNR) catalyzes the de novo synthesis of deoxyribonucleotides in mammals and many other organisms. The RNR subunit R2 contains a dinuclear iron center, which in its diferrous form spontaneously reacts with O2, forming a mu-oxo-bridged diferric cluster and a stable tyrosyl radical. Here, we present the first crystal structures of R2 from mouse with its native dinuclear iron center, both under reducing and oxidizing conditions. In one structure obtained under reducing conditions, the iron-bridging ligand Glu-267 adopts the mu-(eta1,eta2) coordination mode, which has previously been related to O2 activation, and an acetate ion from the soaking solution is observed where O2 has been proposed to bind the iron. The structure of mouse R2 under oxidizing conditions resembles the nonradical diferric R2 from Escherichia coli, with the exception of the coordination of water and Asp-139 to Fe1. There are also additional water molecules near the tyrosyl radical site, as suggested by previous spectroscopic studies. Since no crystal structure of the active radical form has been reported, we propose models for the movement of waters and/or tyrosyl radical site when diferric R2 is oxidized to the radical form, in agreement with our previous ENDOR study. Compared with E. coli R2, two conserved phenylalanine residues in the hydrophobic environment around the diiron center have opposing rotameric conformations, and the carboxylate ligands of the diiron center in mouse R2 appear more flexible. Together, this might contribute to the lower affinity and cooperative binding of iron in mouse R2. Crystal structural studies of changes in the native dinuclear iron center of ribonucleotide reductase protein R2 from mouse.,Strand KR, Karlsen S, Kolberg M, Rohr AK, Gorbitz CH, Andersson KK J Biol Chem. 2004 Nov 5;279(45):46794-801. Epub 2004 Aug 17. PMID:15322079[1] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences
|
| ||||||||||||||||||