1qjg: Difference between revisions
No edit summary |
No edit summary |
||
| (13 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
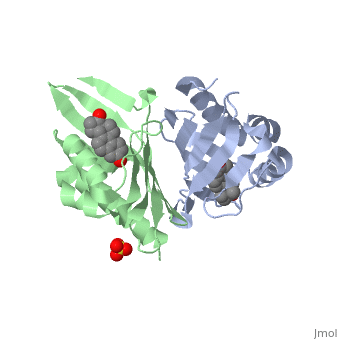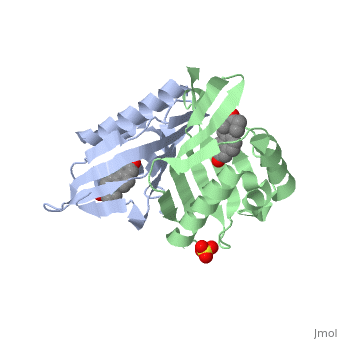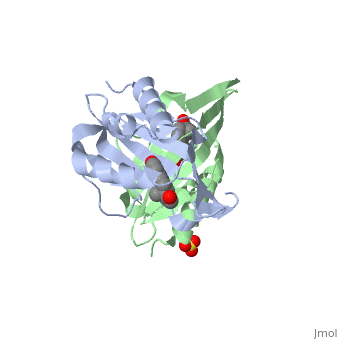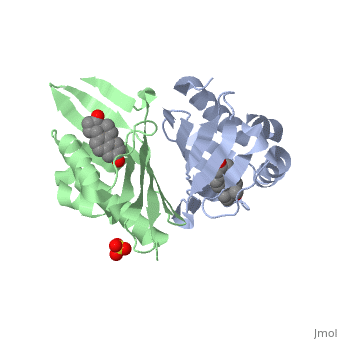
==Crystal structure of delta5-3-ketosteroid isomerase from Pseudomonas testosteroni in complex with equilenin== | |||
<StructureSection load='1qjg' size='340' side='right'caption='[[1qjg]], [[Resolution|resolution]] 2.30Å' scene=''> | |||
== Structural highlights == | |||
<table><tr><td colspan='2'>[[1qjg]] is a 6 chain structure with sequence from [https://en.wikipedia.org/wiki/Comamonas_testosteroni Comamonas testosteroni]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1QJG OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=1QJG FirstGlance]. <br> | |||
</td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 2.3Å</td></tr> | |||
<tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=EQU:EQUILENIN'>EQU</scene>, <scene name='pdbligand=SO4:SULFATE+ION'>SO4</scene></td></tr> | |||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1qjg FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1qjg OCA], [https://pdbe.org/1qjg PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1qjg RCSB], [https://www.ebi.ac.uk/pdbsum/1qjg PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1qjg ProSAT]</span></td></tr> | |||
</table> | |||
== Function == | |||
[https://www.uniprot.org/uniprot/SDIS_COMTE SDIS_COMTE] | |||
== Evolutionary Conservation == | |||
[[Image:Consurf_key_small.gif|200px|right]] | |||
Check<jmol> | |||
<jmolCheckbox> | |||
<scriptWhenChecked>; select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/qj/1qjg_consurf.spt"</scriptWhenChecked> | |||
<scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> | |||
<text>to colour the structure by Evolutionary Conservation</text> | |||
</jmolCheckbox> | |||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1qjg ConSurf]. | |||
<div style="clear:both"></div> | |||
<div style="background-color:#fffaf0;"> | |||
== Publication Abstract from PubMed == | |||
Delta(5)-3-Ketosteroid isomerase from Pseudomonas testosteroni has been intensively studied as a prototype to understand an enzyme-catalyzed allylic isomerization. Asp(38) (pK(a) approximately 4.7) was identified as the general base abstracting the steroid C4beta proton (pK(a) approximately 12.7) to form a dienolate intermediate. A key and common enigmatic issue involved in the proton abstraction is the question of how the energy required for the unfavorable proton transfer can be provided at the active site of the enzyme and/or how the thermodynamic barrier can be drastically reduced. Answering this question has been hindered by the existence of two differently proposed enzyme reaction mechanisms. The 2.26 A crystal structure of the enzyme in complex with a reaction intermediate analogue equilenin reveals clearly that both the Tyr(14) OH and Asp(99) COOH provide direct hydrogen bonds to the oxyanion of equilenin. The result negates the catalytic dyad mechanism in which Asp(99) donates the hydrogen bond to Tyr(14), which in turn is hydrogen bonded to the steroid. A theoretical calculation also favors the doubly hydrogen-bonded system over the dyad system. Proton nuclear magnetic resonance analyses of several mutant enzymes indicate that the Tyr(14) OH forms a low barrier hydrogen bond with the dienolic oxyanion of the intermediate. | |||
Crystal structure of delta(5)-3-ketosteroid isomerase from Pseudomonas testosteroni in complex with equilenin settles the correct hydrogen bonding scheme for transition state stabilization.,Cho HS, Ha NC, Choi G, Kim HJ, Lee D, Oh KS, Kim KS, Lee W, Choi KY, Oh BH J Biol Chem. 1999 Nov 12;274(46):32863-8. PMID:10551849<ref>PMID:10551849</ref> | |||
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | |||
</div> | |||
<div class="pdbe-citations 1qjg" style="background-color:#fffaf0;"></div> | |||
== | ==See Also== | ||
*[[Ketosteroid Isomerase|Ketosteroid Isomerase]] | |||
== References == | |||
<references/> | |||
__TOC__ | |||
</StructureSection> | |||
[[Category: Comamonas testosteroni]] | [[Category: Comamonas testosteroni]] | ||
[[Category: | [[Category: Large Structures]] | ||
[[Category: Cho | [[Category: Cho H-S]] | ||
[[Category: Oh | [[Category: Oh B-H]] | ||
Latest revision as of 15:46, 13 December 2023
Crystal structure of delta5-3-ketosteroid isomerase from Pseudomonas testosteroni in complex with equileninCrystal structure of delta5-3-ketosteroid isomerase from Pseudomonas testosteroni in complex with equilenin
Structural highlights
FunctionEvolutionary Conservation Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. Publication Abstract from PubMedDelta(5)-3-Ketosteroid isomerase from Pseudomonas testosteroni has been intensively studied as a prototype to understand an enzyme-catalyzed allylic isomerization. Asp(38) (pK(a) approximately 4.7) was identified as the general base abstracting the steroid C4beta proton (pK(a) approximately 12.7) to form a dienolate intermediate. A key and common enigmatic issue involved in the proton abstraction is the question of how the energy required for the unfavorable proton transfer can be provided at the active site of the enzyme and/or how the thermodynamic barrier can be drastically reduced. Answering this question has been hindered by the existence of two differently proposed enzyme reaction mechanisms. The 2.26 A crystal structure of the enzyme in complex with a reaction intermediate analogue equilenin reveals clearly that both the Tyr(14) OH and Asp(99) COOH provide direct hydrogen bonds to the oxyanion of equilenin. The result negates the catalytic dyad mechanism in which Asp(99) donates the hydrogen bond to Tyr(14), which in turn is hydrogen bonded to the steroid. A theoretical calculation also favors the doubly hydrogen-bonded system over the dyad system. Proton nuclear magnetic resonance analyses of several mutant enzymes indicate that the Tyr(14) OH forms a low barrier hydrogen bond with the dienolic oxyanion of the intermediate. Crystal structure of delta(5)-3-ketosteroid isomerase from Pseudomonas testosteroni in complex with equilenin settles the correct hydrogen bonding scheme for transition state stabilization.,Cho HS, Ha NC, Choi G, Kim HJ, Lee D, Oh KS, Kim KS, Lee W, Choi KY, Oh BH J Biol Chem. 1999 Nov 12;274(46):32863-8. PMID:10551849[1] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences
|
| ||||||||||||||||||