|
|
| (39 intermediate revisions by one other user not shown) |
| Line 3: |
Line 3: |
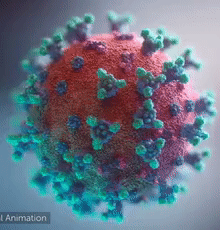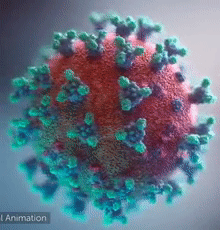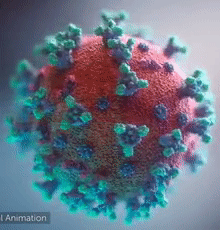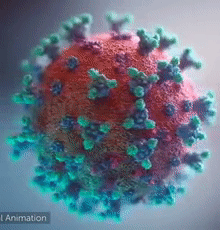
| [[Image:Ezgif.com-video-to-gif 240px 65pc speed.gif|left|240px|thumb|<span style="font-size:100%">SARS-CoV-2 virus. The <span style="color:blue">spikes</span>, that adorn the <span style="color:red">'''virus surface'''</span>, impart a '''''corona''''' like appearance [http://www.drugtargetreview.com/news/57287/3d-visualisation-of-covid-19-surface-released-for-researchers (Fusion Animation)].</span>]] | | [[Image:Ezgif.com-video-to-gif 240px 65pc speed.gif|left|240px|thumb|<span style="font-size:100%">SARS-CoV-2 virus. The <span style="color:blue">spikes</span>, that adorn the <span style="color:red">'''virus surface'''</span>, impart a '''''corona''''' like appearance [http://www.drugtargetreview.com/news/57287/3d-visualisation-of-covid-19-surface-released-for-researchers (Fusion Animation)].</span>]] |
| <span style="font-size:100%"> | | <span style="font-size:100%"> |
| A novel coronavirus SARS-CoV-2, detected in Wuhan, China in 2019, was found to cause severe respiratory illness named '''''COVID-19'''''<ref>[http://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-it Naming the coronavirus disease (COVID-19) and the virus that causes it]</ref>. | | A novel coronavirus SARS-CoV-2, detected in Wuhan, China in 2019, was found to cause severe respiratory illness named '''''COVID-19'''''<ref>[http://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-it Naming the coronavirus disease (COVID-19) and the virus that causes it]</ref> ('''CO'''rona '''VI'''rus '''D'''isease-20'''19'''). |
|
| |
|
| There’s a new [http://go.usa.gov/xw9MV NCBI site] on COVID-19/SARS-CoV-2. It will help bench scientists, bioinformaticians, clinicians & others connect with information they need to study SARS-CoV-2 and end the pandemic. | | There’s a new [http://go.usa.gov/xw9MV NCBI site] on COVID-19/SARS-CoV-2. It will help bench scientists, bioinformaticians, clinicians & others connect with information they need to study SARS-CoV-2 and end the pandemic. |
| Line 9: |
Line 9: |
| A video (by [http://elarasystems.com Elara)] shows how the virus [http://youtu.be/hwVl_-lnoys '''interacts with its human (host) cells'''], via its ''spike'' proteins<ref name="Pillay">PMID:32376714</ref><ref name="McLellan">PMID:32075877</ref><ref name="Veesler">PMID:32155444</ref><ref name="wrobel">PMID: 32647346</ref><ref name="thomas">PMID:12360192</ref><ref name="wang-qi">PMID:32275855</ref>, thus permitting the viral genome to enter its host & begin infection. The spike protein is induced for optimal binding when it is clipped by a protease (see animation on right side) and more details at '''[[SARS-CoV-2_protein_S_activation_by_furin]]'''. Subsequently, it undergoes a dramatic transformation to induce membrane fusion and infection, explained with a morph animation at '''[[SARS-CoV-2 spike protein fusion transformation]]'''. | | A video (by [http://elarasystems.com Elara)] shows how the virus [http://youtu.be/hwVl_-lnoys '''interacts with its human (host) cells'''], via its ''spike'' proteins<ref name="Pillay">PMID:32376714</ref><ref name="McLellan">PMID:32075877</ref><ref name="Veesler">PMID:32155444</ref><ref name="wrobel">PMID: 32647346</ref><ref name="thomas">PMID:12360192</ref><ref name="wang-qi">PMID:32275855</ref>, thus permitting the viral genome to enter its host & begin infection. The spike protein is induced for optimal binding when it is clipped by a protease (see animation on right side) and more details at '''[[SARS-CoV-2_protein_S_activation_by_furin]]'''. Subsequently, it undergoes a dramatic transformation to induce membrane fusion and infection, explained with a morph animation at '''[[SARS-CoV-2 spike protein fusion transformation]]'''. |
| </span> | | </span> |
|
| |
|
| |
|
| |
|
| |
|
| |
|
|
| |
|
| == Useful sites on COVID-19 == | | == Useful sites on COVID-19 == |
| * A plot of the number of COVID-19 deaths per million people as reported at the [https://ourworldindata.org/grapher/weekly-covid-deaths-per-million-people|Johns Hopkins Web Site], Surprisingly, Israel and USA have the highest numbers.<br /> | | * A plot of the Biweekly change in confirmed COVID-19 cases throughout the world |
| [[Image:JHU Weekly confirmed COVID-19 deaths per Million people.jpg|350px|left|thumb|COVID-19 deaths per million peopled)<ref name="JHU Web Site"> JHU COVID-19 Web Site https://ourworldindata.org/grapher/weekly-covid-deaths-per-million-people </ref>]].<br />
| | <imagemap> |
| | | Image:Biweekly change in confirmed COVID-19 cases.jpg|500px |
| * A series of fascinating '''[[COVID-19 Videos]]'''.
| | default [https://ourworldindata.org/grapher/biweekly-growth-covid-cases Our World site with the most recent data] |
| | | </imagemap> |
| * '''[[COVID-19 AlphaFold2 Models]]''' that have been of structures of SARS CoV-2 proteins whose structure have not yet been experimentally determined.
| | Clicking the above image goes to the [https://ourworldindata.org/grapher/biweekly-growth-covid-cases Our World in Data] site with the most recent data. |
| | |
| * Up to date statistics on [http://www.worldometers.info/coronavirus/coronavirus-cases/ Coronavirus cases world-wide at worldometer]
| |
|
| |
|
| * A summary of key findings about [http://www.cdc.gov/coronavirus/2019-nCoV/lab/index.html COVID-19] can be found at [http://www.cdc.gov/ the '''CDC''']. | | * A summary of key findings about [http://www.cdc.gov/coronavirus/2019-nCoV/lab/index.html COVID-19] can be found at [http://www.cdc.gov/ the '''CDC''']. |
|
| |
|
| * '''Coronavirus Evolved Naturally''', and ‘Is '''Not''' a Laboratory Construct,’ in a study in Nature Med by Anderson and colleagues <ref>Andersen, et al. The proximal origin of SARS-CoV-2: Nature Med (in press) 2020 [http://dx.doi.org/10.1038/s41591-020-0820-9 http://dx.doi.org/10.1038/s41591-020-0820-9]]</ref>. | | * A series of fascinating '''[[COVID-19 Videos]]'''. |
| == Potential treatments for COVID-19 ==
| |
| * [http://www.dropbox.com/s/tb5nvaq7f6o38kt/COVID-19%204.12.20.pdf?dl=0 '''COVID-19: What are our therapeutic options?'''] - Slides courtesy of [http://www.utsouthwestern.edu/labs/jain-mamta/ '''Prof. Mamta K. Jain, MD, MPH'''], [http://www.utsouthwestern.edu/ UT Southwestern Medical Center].
| |
|
| |
|
| * An int'l group of scientists (UK, Israel & USA) are trying to combat COVID-19 via a massive crystal-based fragment screen. They determined 3D structures of [http://www.diamond.ac.uk/covid-19/for-scientists/Main-protease-structure-and-XChem/Downloads.html over '''90 fragments''' with '''66 in the active site''']. Details are available [http://www.diamond.ac.uk/covid-19/for-scientists/Main-protease-structure-and-XChem.html at the '''Diamond Light Source''']. Contributions are welcome in many different forms [http://covid.postera.ai/covid '''PostEra''']. | | * '''[[Potential treatments for COVID-19]]'''. |
|
| |
|
| * 31-Mar-2020 ''NY Times'' [http://www.nytimes.com/2020/03/31/health/cdc-masks-coronavirus.html C.D.C. Weighs Advising Everyone to Wear a Mask]. | | * '''[[SARS-CoV-2 virus proteins]]''' - Status of all 3D models of SARS-CoV-2 Virus protein structures. |
|
| |
|
| == 3D structural studies on SARS-CoV-2 virus==
| | * '''[[COVID-19 AlphaFold2 Models]]''' that have been of structures of SARS CoV-2 proteins whose structure have not yet been experimentally determined. |
| {{Template:COVID Validation}}
| |
| | |
| * A team of UK & Israeli scientists determined the 3D structures of [http://www.diamond.ac.uk/covid-19/for-scientists/Main-protease-structure-and-XChem/Downloads.html '''over 90 fragments, 66 of which are in the active site'']. All the experimental details and results are available [http://www.diamond.ac.uk/covid-19/for-scientists/Main-protease-structure-and-XChem.html '''online at the Diamond Light Source''']. | |
| | |
| * A team of Chinese scientists determined, by Cryo-EM, the''' coronavirus spike receptor-binding domain complexed with its receptor ACE2 PDB-ID''' [[6lzg]] (To be published).
| |
| | |
| * A team of US and Chinese scientists determined the crystal structure of '''2019-nCoV spike receptor-binding domain bound with ACE2''' [[6m0j]]
| |
| | |
| * A team of US scientists determined, by Cryo-EM, the '''structure of the SARS-CoV-2 spike glycoprotein (open & closed states)''' <ref>PMID:32155444</ref>, PDB-ID [[6vxx]] and [[6vyb]]. See an [[SARS-CoV-2 protein S priming by furin|animation of this transition]].
| |
| | |
| * Crystal structure of SARS-CoV-2 receptor binding domain in complex with human antibody CR3022 [[6w41]]
| |
| | |
| * Crystal Structure of the methyltransferase-stimulatory factor complex of NSP16 and NSP10 from SARS CoV-2 [[6w61]] (To be published).
| |
| | |
| * Crystal Structure of ADP ribose phosphatase of NSP3 from SARS CoV-2 in complex with AMP [[6w6y]] (To be published).
| |
| | |
| * Structure of NSP10 - NSP16 Complex from SARS-CoV-2 [[6w75]] (To be published).
| |
| | |
| * Crystal structure of SARS-CoV-2 nucleocapsid protein N-terminal RNA binding domain [[6m3m]] (To be published).
| |
| | |
| * Crystal structure of Nsp9 RNA binding protein of SARS CoV-2 [[6w4b]] (To be published).
| |
| | |
| * Crystal Structure of NSP16 - NSP10 Complex from SARS-CoV-2 [[6w4h]] (To be published).
| |
| | |
| * A Cryo-EM study by Zhou & colleagues on the structural basis for the '''recognition of the SARS-CoV-2 by full-length human ACE2''' gives insights to the molecular basis for coronavirus recognition and infection<ref>PMID:32132184</ref> [[6m17]].
| |
| | |
| * Crystal structure of RNA binding domain of nucleocapsid phosphoprotein from SARS coronavirus 2 [[6vyo]] (To be published).
| |
| | |
| * Crystal structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with a Citrate [[6w01]] (To be published).
| |
| | |
| * Crystal structure of ADP ribose phosphatase of NSP3 from SARS CoV-2 in the complex with ADP ribose [[6w02]] (To be published).
| |
| | |
| * Crystal structure of 2019-nCoV chimeric receptor-binding domain complexed with its receptor human ACE2 [[6vw1]]
| |
| | |
| * Crystal structure of NSP15 Endoribonuclease from SARS CoV-2 [[6vww]] (To be published).
| |
| | |
| * Crystal Structure of ADP ribose phosphatase of NSP3 from SARS CoV-2 [[6vxs]] (To be published).
| |
| | |
| * Crystal structure of the 2019-nCoV HR2 Domain [[6lvn]] (To be published).
| |
| | |
| * Crystal structure of post fusion core of 2019-nCoV S2 subunit [[6lxt]].
| |
| | |
| * Crystal structure of '''SARS-CoV-2 main protease''' provides a basis for design of improved α-ketoamide inhibitors, from the Hilgenfeld lab<ref>PMID:32198291</ref>, Apo Struture: PDB-ID [[6y2e]], and complexes with inhibitors: PDB-ID [[6y2f ]] and [[6y2g]].
| |
| | |
| * 3D Structure of '''RNA-dependent RNA polymerase from COVID-19''', a '''major antiviral drug target''' from the Rao lab in Beijing<ref> Gao, et al. Structure of RNA-dependent RNA polymerase from 2019-nCoV, a major antiviral drug target: bioRxiv (online) 2020 [http://doi.org/10.1101/2020.03.16.993386 http://doi.org/10.1101/2020.03.16.993386]</ref>.
| |
| | |
| * Crystal structure of the '''Mpro from COVID-19''' and '''discovery of inhibitors''' in a study by scientists from Shanghai & Beijing<ref>PMID: 32272481</ref>, PDB-ID [[6lu7]].
| |
| | |
| * Crystal structure of '''Nsp15 endoribonuclease NendoU from SARS-CoV-2''' in a study by scientists from USA<ref> Kim, et al. Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2: bioRxiv (online) 2020 [http://doi.org/10.1101/2020.03.02.968388 http://doi.org/10.1101/2020.03.02.968388]</ref>, PDB-ID [[6w01]].
| |
| | |
| * '''The CoV spike (S) glycoprotein is a key target for vaccines, therapeutic antibodies, and diagnostics'''. A study by McLellan and colleagues in "Science" on the Cryo-EM structure of the COVID-19 spike protein. This structure should greatly aid in the rapid development and evaluation of medical countermeasures to address the ongoing public health crisis<ref name="McLellan" />, PDB-ID [[6vsb]].
| |
| | |
| * [http://github.com/thorn-lab/coronavirus_structural_task_force Public resource for the structures from beta-coronavirus with a focus on SARS-CoV and SARS-CoV-2]
| |
|
| |
|
| == SARS-CoV-2 virus proteins ==
| | * '''[[3D structural studies on SARS-CoV-2]]''' - Pointers to research on 3D structural studies on SARS-CoV-2 Virus proteins. |
| {{Template:COVID Validation}}
| |
|
| |
|
| The genome of the SARS-CoV-2 virus codes for 28 proteins:
| |
| Out of those, 19 have already been characterized structurally.
| |
| <br>
| |
| Details of the '''3D structure & function''' of the key proteins & RNA inside the virus can be seen in the NY Times [http://www.nytimes.com/interactive/2020/04/03/science/coronavirus-genome-bad-news-wrapped-in-protein.html '''''Bad News Wrapped in Protein: Inside the Coronavirus Genome'''''].
| |
|
| |
|
| * [[SARS-CoV-2 Coronavirus Main Protease| Main protease]]: it is a cysteine protease that is essential for the viral life cycle.
| |
| * [[SARS-CoV-2_protein_NSP1|NSP1]]: ''Host translation inhibitor nsp1''. Inhibits host translation by interacting with the 40S ribosomal subunit.
| |
| * [[SARS-CoV-2_protein_NSP2|NSP2]]: ''Non-structural protein 2''.
| |
| * [[SARS-CoV-2 enzyme Papain-like|Papain-like proteinase]]
| |
| * [[SARS-CoV-2_protein_NSP4|NSP4]]: ''Non-structural protein 4''.
| |
| * [[SARS-CoV-2 enzyme 3CL-PRO|Proteinase 3CL-PRO]]
| |
| * '''''AlphaFold2 Theoretical Model''''' [[SARS-CoV-2_protein_NSP6|NSP6]]: ''Non-structural protein 6''.
| |
| * [[SARS-CoV-2_protein_NSP7|NSP7]]: ''Non-structural protein 7''.
| |
| * [[SARS-CoV-2_protein_NSP8|NSP8]]: ''Non-structural protein 8''.
| |
| * [[SARS-CoV-2_protein_NSP9|NSP9]]: ''Non-structural protein 9''.
| |
| * [[SARS-CoV-2_protein_NSP10|NSP10]]: ''Non-structural protein 10''.
| |
| * [[SARS-CoV-2 enzyme RdRp|RNA-directed RNA polymerase (RdRp)]]
| |
| * [[SARS-CoV-2 enzyme Hel|Helicase (Hel)]]: Multi-functional protein with a zinc-binding domain in N-terminus
| |
| * [[SARS-CoV-2 enzyme ExoN|Guanine-N7 methyltransferase (ExoN)]]
| |
| * [[SARS-CoV-2 enzyme NendoU|Uridylate-specific endoribonuclease (NendoU)]]
| |
| * [[SARS-CoV-2 enzyme 2'-O-MT|2'-O-methyltransferase (2'-O-MT)]]: Methyltransferase that mediates mRNA cap 2'-O-ribose methylation to the 5'-cap structure of viral mRNAs.
| |
| * [[SARS-CoV-2_protein_S|Surface glycoprotein (S)]] and [[SARS-CoV-2 protein S priming by furin]]
| |
| * [[SARS-CoV-2 protein ORF3a|ORF3a]]: Forms homotetrameric potassium sensitive ion channels (viroporin) and may modulate virus release.
| |
| * [[SARS-CoV-2_protein_E|Protein E]]: Plays a central role in virus morphogenesis and assembly.
| |
| * '''''AlphaFold2 Theoretical Model''''' [[SARS-CoV-2_protein_M|Protein M]]: Component of the viral envelope.
| |
| * [[SARS-CoV-2 protein ORF6|ORF6]]: Could be a determinant of virus virulence.
| |
| * [[SARS-CoV-2 protein ORF7a|ORF7a]]: Non-structural protein which is dispensable for virus replication in cell culture.
| |
| * [[SARS-CoV-2 protein ORF8|ORF8]]: Open Reading Frame 8.
| |
| * '''''AlphaFold2 Theoretical Model''''' [[SARS-CoV-2_protein_N|Protein N]]: Packages the positive strand viral genome RNA into a helical ribonucleocapsid (RNP).
| |
| * '''''AlphaFold2 Theoretical Model''''' [[SARS-CoV-2 protein ORF10|ORF10]]: It is currently unclear whether this region translates into a functional protein.
| |
|
| |
|
| == References == | | == References == |