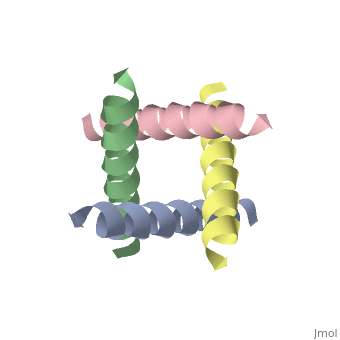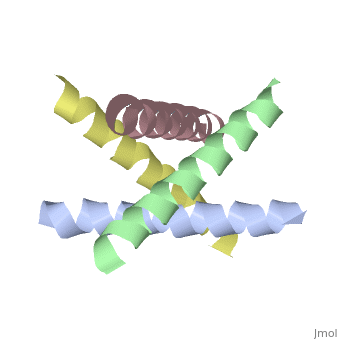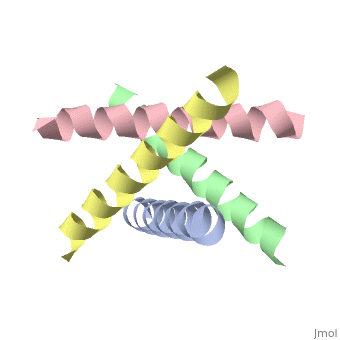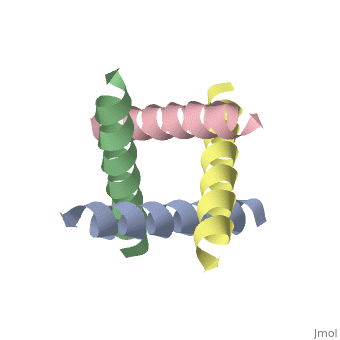1nyj: Difference between revisions
New page: left|200px<br /><applet load="1nyj" size="450" color="white" frame="true" align="right" spinBox="true" caption="1nyj" /> '''The closed state structure of M2 protein H+ ... |
No edit summary |
||
| (18 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
== | ==The closed state structure of M2 protein H+ channel by solid state NMR spectroscopy== | ||
An interhelical distance has been precisely measured by REDOR solid-state | <StructureSection load='1nyj' size='340' side='right'caption='[[1nyj]]' scene=''> | ||
== Structural highlights == | |||
<table><tr><td colspan='2'>[[1nyj]] is a 4 chain structure with sequence from [https://en.wikipedia.org/wiki/Influenza_A_virus_(A/USSR/90/1977(H1N1)) Influenza A virus (A/USSR/90/1977(H1N1))]. Full experimental information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1NYJ OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=1NYJ FirstGlance]. <br> | |||
</td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">Solid-state NMR</td></tr> | |||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1nyj FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1nyj OCA], [https://pdbe.org/1nyj PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1nyj RCSB], [https://www.ebi.ac.uk/pdbsum/1nyj PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1nyj ProSAT]</span></td></tr> | |||
</table> | |||
== Function == | |||
[https://www.uniprot.org/uniprot/M2_I77AB M2_I77AB] Forms a proton-selective ion channel that is necessary for the efficient release of the viral genome during virus entry. After attaching to the cell surface, the virion enters the cell by endocytosis. Acidification of the endosome triggers M2 ion channel activity. The influx of protons into virion interior is believed to disrupt interactions between the viral ribonucleoprotein (RNP), matrix protein 1 (M1), and lipid bilayers, thereby freeing the viral genome from interaction with viral proteins and enabling RNA segments to migrate to the host cell nucleus, where influenza virus RNA transcription and replication occur. Also plays a role in viral proteins secretory pathway. Elevates the intravesicular pH of normally acidic compartments, such as trans-Golgi network, preventing newly formed hemagglutinin from premature switching to the fusion-active conformation (By similarity). | |||
<div style="background-color:#fffaf0;"> | |||
== Publication Abstract from PubMed == | |||
An interhelical distance has been precisely measured by REDOR solid-state NMR spectroscopy in the transmembrane tetrameric bundle of M2-TMP, from the M2 proton channel of the influenza A viral coat. The high-resolution structure of the helical backbone has been determined using orientational restraints from uniformly aligned peptide preparations in hydrated dimyristoylphosphatidylcholine bilayers. Here, the distance between (15)N(pi) labeled His37 and (13)C(gamma) labeled Trp41 is determined to be less than 3.9 A. Such a short distance, in combination with the known tilt and rotational orientation of the individual helices, permits not only a determination of which specific side chain pairings give rise to the interaction, but also the side chain torsion angles and restraints for the tetrameric bundle can also be characterized. The resulting proton channel structure is validated in a variety of ways. Both histidine and tryptophan side chains are oriented in toward the pore where they can play a significant functional role. The channel appears to be closed by the proximity of the four indoles consistent with electrophysiology and mutagenesis studies of the intact protein at pH 7.0 and above. The pore maintains its integrity to the N terminal side of the membrane, and at the same time, a cavity is generated that appears adequate for binding amantadine. Finally, the observation of a 2 kHz coupling in the PISEMA spectrum of (15)N(pi)His37 validates the orientation of the His37 side chain based on the observed REDOR distance. | |||
The closed state of a H+ channel helical bundle combining precise orientational and distance restraints from solid state NMR.,Nishimura K, Kim S, Zhang L, Cross TA Biochemistry. 2002 Nov 5;41(44):13170-7. PMID:12403618<ref>PMID:12403618</ref> | |||
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | |||
</div> | |||
<div class="pdbe-citations 1nyj" style="background-color:#fffaf0;"></div> | |||
==See Also== | |||
*[[Ion channels 3D structures|Ion channels 3D structures]] | |||
*[[M2 protein|M2 protein]] | |||
== References == | |||
<references/> | |||
__TOC__ | |||
</StructureSection> | |||
[[Category: Large Structures]] | |||
[[Category: Cross TA]] | |||
[[Category: Kim S]] | |||
[[Category: Nishimura K]] | |||
[[Category: Zhang L]] | |||
Latest revision as of 21:53, 29 November 2023
The closed state structure of M2 protein H+ channel by solid state NMR spectroscopyThe closed state structure of M2 protein H+ channel by solid state NMR spectroscopy
Structural highlights
FunctionM2_I77AB Forms a proton-selective ion channel that is necessary for the efficient release of the viral genome during virus entry. After attaching to the cell surface, the virion enters the cell by endocytosis. Acidification of the endosome triggers M2 ion channel activity. The influx of protons into virion interior is believed to disrupt interactions between the viral ribonucleoprotein (RNP), matrix protein 1 (M1), and lipid bilayers, thereby freeing the viral genome from interaction with viral proteins and enabling RNA segments to migrate to the host cell nucleus, where influenza virus RNA transcription and replication occur. Also plays a role in viral proteins secretory pathway. Elevates the intravesicular pH of normally acidic compartments, such as trans-Golgi network, preventing newly formed hemagglutinin from premature switching to the fusion-active conformation (By similarity). Publication Abstract from PubMedAn interhelical distance has been precisely measured by REDOR solid-state NMR spectroscopy in the transmembrane tetrameric bundle of M2-TMP, from the M2 proton channel of the influenza A viral coat. The high-resolution structure of the helical backbone has been determined using orientational restraints from uniformly aligned peptide preparations in hydrated dimyristoylphosphatidylcholine bilayers. Here, the distance between (15)N(pi) labeled His37 and (13)C(gamma) labeled Trp41 is determined to be less than 3.9 A. Such a short distance, in combination with the known tilt and rotational orientation of the individual helices, permits not only a determination of which specific side chain pairings give rise to the interaction, but also the side chain torsion angles and restraints for the tetrameric bundle can also be characterized. The resulting proton channel structure is validated in a variety of ways. Both histidine and tryptophan side chains are oriented in toward the pore where they can play a significant functional role. The channel appears to be closed by the proximity of the four indoles consistent with electrophysiology and mutagenesis studies of the intact protein at pH 7.0 and above. The pore maintains its integrity to the N terminal side of the membrane, and at the same time, a cavity is generated that appears adequate for binding amantadine. Finally, the observation of a 2 kHz coupling in the PISEMA spectrum of (15)N(pi)His37 validates the orientation of the His37 side chain based on the observed REDOR distance. The closed state of a H+ channel helical bundle combining precise orientational and distance restraints from solid state NMR.,Nishimura K, Kim S, Zhang L, Cross TA Biochemistry. 2002 Nov 5;41(44):13170-7. PMID:12403618[1] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences |
| ||||||||||||||||