FirstGlance/Evaluating Protein Crosslinks: Difference between revisions
Eric Martz (talk | contribs) |
Eric Martz (talk | contribs) |
||
| (41 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
FirstGlance in Jmol<!--, available from [http://firstglance.jmol.org firstglance.jmol.org],--> is the easiest-to-use tool for visualizing, understanding, and sharing protein molecular structure. It offers guided exploration with extensive explanations and examples, yet provides ample power for advanced users. With a few mouse clicks, any molecular scene can be saved as a slide presentation-ready animation (GIF file) -- see examples below, and [http://tinyurl.com/movingmolecules tinyurl.com/movingmolecules]. | FirstGlance in Jmol<!--, available from [http://firstglance.jmol.org firstglance.jmol.org],--> is the easiest-to-use tool for visualizing, understanding, and sharing protein molecular structure. It offers guided exploration with extensive explanations and examples, yet provides ample power for advanced users. With a few mouse clicks, any molecular scene can be saved as a slide presentation-ready animation (GIF file) -- see examples below, and [http://tinyurl.com/movingmolecules tinyurl.com/movingmolecules]. | ||
Below are step-by-step instructions for evaluating [[protein crosslinks]] | Below are step-by-step instructions for evaluating putative [[protein crosslinks]] reported by FirstGlance in Jmol. Genuine crosslink bonds are illustrated, along with [[clashes]] that are irregularities in the model, not crosslink bonds. | ||
==Instructions for an Isopeptide Bond== | ==Instructions for an Isopeptide Bond== | ||
1. | 1. Go to [http://firstglance.jmol.org FirstGlance in Jmol] (right click link, open in new tab), and enter [[PDB code]] <b>2xi9</b>. | ||
<br> | <br> | ||
<table class="wikitable"><tr><td>[[Image:Fgij-xl1.png]]</td></tr></table> | <table class="wikitable"><tr><td>[[Image:Fgij-xl1.png]]</td></tr></table> | ||
| Line 44: | Line 40: | ||
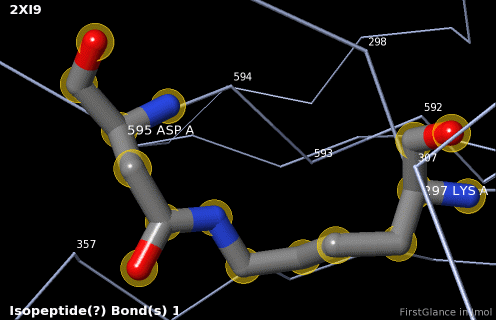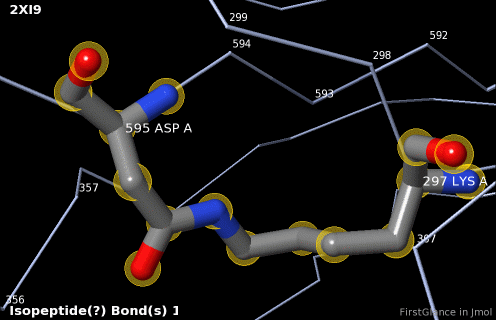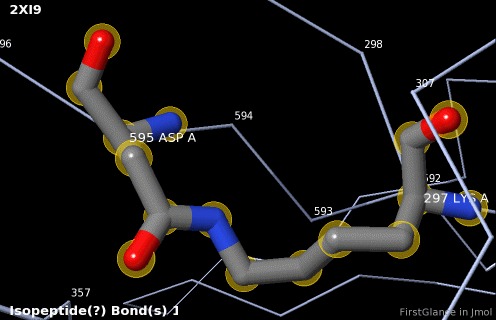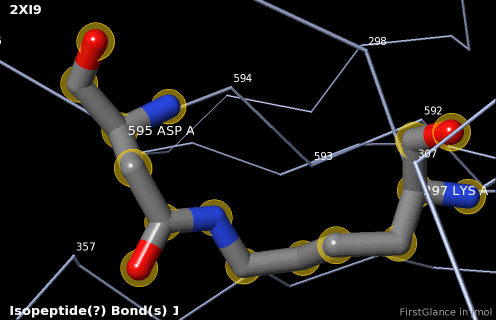
6. Finally, click on the link to show the ''Electron Density Map''. At the very good resolution of 1.9 Å, every atom in this pair of residues has clear electron density. There is no ambiguity about where these atoms belong in the model. Therefore, the electron density map supports the presence of this isopeptide bond. | 6. Finally, click on the link to show the ''Electron Density Map''. At the very good [[resolution]] of 1.9 Å, every atom in this pair of residues has clear electron density. There is no ambiguity about where these atoms belong in the model. Therefore, the electron density map supports the presence of this isopeptide bond. | ||
<br> | <br> | ||
<table class="wikitable"><tr><td>[[Image:Fgij-xl8.png]]</td><td>[[Image:Fgij-xl8.gif]]</td></tr></table> | <table class="wikitable"><tr><td>[[Image:Fgij-xl8.png]]</td><td>[[Image:Fgij-xl8.gif]]</td></tr></table> | ||
===Conservation of Crosslinking Residues=== | |||
If the PDB file is pre-processed by [[ConSurf/Index|ConSurf]], after displaying the crosslinked residues, their levels of evolutionary conservation will be shown by checking ''ConSurf Colors'' (in the middle-left of FirstGlance). | |||
'''For more complete instructions and illustrations, see [[Interpreting ConSurf Results]]'''. | |||
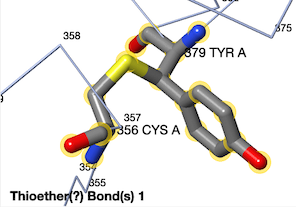
For example, below is a thioether covalent crosslink between Cys356 and Tyr379 in catalase [[1sy7]]. (The multiple sequence alignment included 150 sequences sampled from 22,855 unique similar sequences. The [[Interpreting_ConSurf_Results#Average_Pairwise_Distance|average pairwise distance]] was 0.96.) | |||
<table class="wikitable"><tr><td> | |||
[[Image:1sy7-fg-thioether.png]] | |||
</td><td width="350"> | |||
[[Image:1sy7-fg-consurf-thioether.png]] | |||
</td></tr><tr><td> | |||
Thioether crosslink in catalase [[1sy7]]. | |||
<br> | |||
<span style="font-size:160%;"> | |||
{{Template:ColorKey_Element_C}} | |||
{{Template:ColorKey_Element_N}} | |||
{{Template:ColorKey_Element_O}} | |||
{{Template:ColorKey_Element_S}} | |||
</span> | |||
</td><td> | |||
Crosslinking residues: | |||
<br> | |||
{{Template:ColorKey_ConSurf_NoYellow_NoGray}} | |||
<br> | |||
Heme: | |||
<span style="font-size:160%;"> | |||
{{Template:ColorKey_Element_C}} | |||
{{Template:ColorKey_Element_N}} | |||
{{Template:ColorKey_Element_O}} | |||
{{Template:ColorKey_Element_Fe}} | |||
</span> | |||
<br> | |||
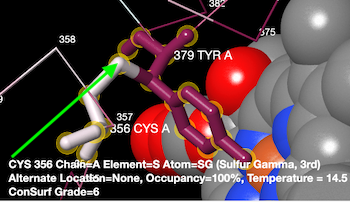
Clicking the Cys sulfur atom (green arrow) produced the report at the bottom. | |||
</td></tr></table> | |||
Additional crosslink examples showing conservation: | |||
*[[FirstGlance/Visualizing_Conservation#Conservation_of_Protein_Crosslinks|An isopeptide crosslink in polyubiquitin]]. | |||
*[[Interpreting_ConSurf_Results#More_Sequences_Needed|An isopeptide in crosslinked lamprey fibrin]]. | |||
==Verified Crosslink Examples== | ==Verified Crosslink Examples== | ||
| Line 55: | Line 90: | ||
==Clashes== | ==Clashes== | ||
FirstGlance suggests the presence of a "putative crosslink" based solely on the distances between relevant atoms (click the link '''Detailed Methods''' in FirstGlance). Sometimes, instead of a real covalent bond crosslink, the proximity of the relevant atoms is due to errors in the atomic model that produce [[clashes]]. | FirstGlance suggests the presence of a "putative crosslink" based solely on the distances between relevant atoms (click the link '''Detailed Methods''' in FirstGlance). Sometimes, instead of a real covalent bond crosslink, the proximity of the relevant atoms is due to [[alternate locations]] or small errors in the atomic model that produce [[clashes]]. | ||
For each putative crosslink suggested by FirstGlance, it is the responsibility of the user to evaluate whether it is a real covalent crosslink, or a clash. FirstGlance makes this responsibility clear (see the bottom paragraph in the snapshot under step #4 above). | For each putative crosslink suggested by FirstGlance, it is the responsibility of the user to evaluate whether it is a real covalent crosslink, or a clash. FirstGlance makes this responsibility clear (see the bottom paragraph in the snapshot under step #4 above). | ||
| Line 61: | Line 96: | ||
===Bonds=== | ===Bonds=== | ||
It is important not to take the bonds displayed by FirstGlance/Jmol literally. [[Atomic coordinate files]] from the [[wwPDB]] do not specify most bonds. Rather, Jmol draws | It is important not to take the bonds displayed by FirstGlance/Jmol literally. [[Atomic coordinate files]] from the [[wwPDB]] do not specify most covalent bonds. Rather, Jmol draws covalent "bond" sticks between atoms wherever two atoms are sufficiently close. In the case of carbon, nitrogen and oxygen, this is wherever a pair of those atoms is separated by ≤1.81 Å (for actual bond lengths, see footnote<ref name="bondlengths">Actual covalent bond lengths are approximately C-C: 1.4 Å; C-N: 1.4 Å; C-O: 1.4 Å; C=O: 1.2 Å; C-S: 1.7 Å ([https://en.wikipedia.org/wiki/Covalent_radius Covalent radii in Wikipedia]).</ref>). In perfectly accurate models, this works correctly, but the errors in [[empirical models]] sometimes lead to bonds being drawn incorrectly. Thus, Jmol may show bond sticks where the reality is a clash, not a bond; and Jmol may fail to show bond sticks between atoms that are believed to be covalently bonded, but are too far apart in the model. | ||
== | ==Case Illustrations== | ||
For each type of crosslink, FirstGlance gives one or more examples of clashes just above the list of verified crosslink examples, explaining the evidence for a clash. Below are a few specific cases. | For each type of crosslink, FirstGlance gives one or more examples of clashes just above the list of verified crosslink examples, explaining the evidence for a clash. Below are a few specific cases. | ||
===His-Tyr=== | |||
====His-Tyr Genuine Crosslink==== | ====His-Tyr Genuine Crosslink==== | ||
| Line 96: | Line 133: | ||
[[Image:Fgij-xl10-histyr-clash.png]] | [[Image:Fgij-xl10-histyr-clash.png]] | ||
</td></tr></table> | </td></tr></table> | ||
[[4rea]] also has a clash suggested as a possible isopeptide bond by FirstGlance. Both that, and the His-Tyr clashes are removed by [[PDB REDO]]. | |||
===Thioesters=== | |||
====Thioester Genuine Crosslink==== | ====Thioester Genuine Crosslink==== | ||
| Line 108: | Line 149: | ||
</span> | </span> | ||
</td></tr></table> | </td></tr></table> | ||
[[2xi9]] has 2 copies of a bacterial pilus adhesin in its [[asymmetric unit]]. FirstGlance reports a putative thioester crosslink in each copy of the protein. The relevant residues are modeled as a thiester bond with correct geometry. The C-S distances in the model are 1.6 Å, in good agreement with the typical bond length of 1.7 Å<ref name="bondlengths" />. The electron density map shows clear density for the thioester-bonded atoms. The difference map (not shown) raises no alarms. The abstract of the publication highlights this thioester bond. | [[2xi9]] has 2 copies of a bacterial pilus adhesin in its [[asymmetric unit]]. FirstGlance reports a putative [[Thioester protein crosslinks|thioester crosslink]] in each copy of the protein. The relevant residues are modeled as a thiester bond with correct geometry. The C-S distances in the model are 1.6 Å, in good agreement with the typical bond length of 1.7 Å<ref name="bondlengths" />. The electron density map shows clear density for the thioester-bonded atoms. The difference map (not shown) raises no alarms. The abstract of the publication highlights this thioester bond. | ||
===="Thioester" Clash==== | ===="Thioester" Clash==== | ||
| Line 128: | Line 169: | ||
</td></tr></table> | </td></tr></table> | ||
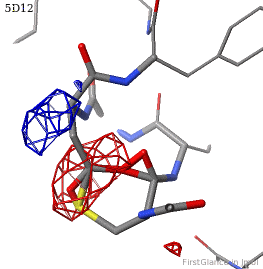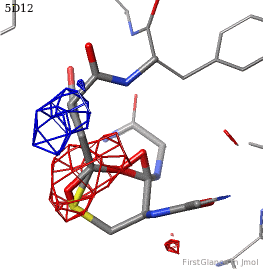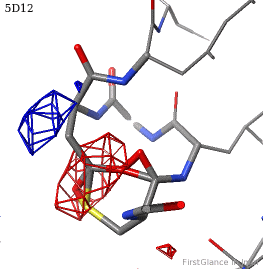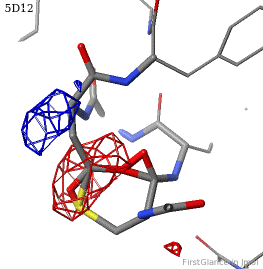
[[5d12]] has two copies of a kinase domain in its [[asymmetric unit]]. FirstGlance reports a possible thioester crosslink | [[5d12]] has two copies of a kinase domain in its [[asymmetric unit]]. FirstGlance reports a possible thioester crosslink only in chain B. The C-S distance is 1.76 Å, quite reasonable. However, the residues are not modeled as a thiester bond, and Jmol draws many spurious "bonds". The sidechain of Asp348 passes through a gap in the all-features electron density (<span style="color:#00a000;font-weight:bold;">green isomesh</span>, above left). The difference electron density map (above right) reveals insufficient density to account for the position of the Asp sidechain (<span style="color:#b00000;font-weight:bold;">dark red isomesh</span>), while suggesting an alternative position for those sidechain atoms (<span style="color:#0000ff;font-weight:bold;">blue isomesh</span>). In conclusion, this is a clash, not a thioester crosslink bond. | ||
After [[PDB REDO]] re-refines and rebuilds 5d12, this clash is absent. | |||
===Lys-Cys NOS=== | |||
====Lys-Cys NOS Genuine Crosslinks==== | ====Lys-Cys NOS Genuine Crosslinks==== | ||
| Line 159: | Line 204: | ||
[[7ns5]] is a homo-tetramer. FirstGlance suggests a possible Lys-Cys NOS crosslink in chain B. The fact that FirstGlance suggests no such crosslinks in the other 3 chains is a hint that the proximity of the sidechain nitrogen of Lys167 to the sulfur in Cys165 in chain B is a clash. The atomic model shows an improbable N-S bond between the sidechains. Further, electron density (above left) is missing for the last 3 atoms of the lysine sidechain, consistent with it being misplaced (<span style="color:#00a000;font-weight:bold;">green isomesh</span>). The difference electron density map (above right) reveals that there is no electron density consistent with the lysine sidechain nitrogen being located where it was placed in the model (<span style="color:#b00000;font-weight:bold;">dark red isomesh</span>). In conclusion, this is a clash in chain B, not an NOS crosslink. | [[7ns5]] is a homo-tetramer. FirstGlance suggests a possible Lys-Cys NOS crosslink in chain B. The fact that FirstGlance suggests no such crosslinks in the other 3 chains is a hint that the proximity of the sidechain nitrogen of Lys167 to the sulfur in Cys165 in chain B is a clash. The atomic model shows an improbable N-S bond between the sidechains. Further, electron density (above left) is missing for the last 3 atoms of the lysine sidechain, consistent with it being misplaced (<span style="color:#00a000;font-weight:bold;">green isomesh</span>). The difference electron density map (above right) reveals that there is no electron density consistent with the lysine sidechain nitrogen being located where it was placed in the model (<span style="color:#b00000;font-weight:bold;">dark red isomesh</span>). In conclusion, this is a clash in chain B, not an NOS crosslink. | ||
After [[PDB REDO]] re-refines and rebuilds 7ns5, this clash is absent. | |||
==See Also== | |||
*[[FirstGlance/Index]]: FirstGlance resources within and external to Proteopedia. | |||
==Notes and References== | ==Notes and References== | ||
<references /> | <references /> | ||
Latest revision as of 20:26, 9 November 2023
FirstGlance in Jmol is the easiest-to-use tool for visualizing, understanding, and sharing protein molecular structure. It offers guided exploration with extensive explanations and examples, yet provides ample power for advanced users. With a few mouse clicks, any molecular scene can be saved as a slide presentation-ready animation (GIF file) -- see examples below, and tinyurl.com/movingmolecules.
Below are step-by-step instructions for evaluating putative protein crosslinks reported by FirstGlance in Jmol. Genuine crosslink bonds are illustrated, along with clashes that are irregularities in the model, not crosslink bonds.
Instructions for an Isopeptide BondInstructions for an Isopeptide Bond
1. Go to FirstGlance in Jmol (right click link, open in new tab), and enter PDB code 2xi9.
 |
2. FirstGlance automatically alerts you that unusual protein crosslinks are present. The alert is different according to whether FirstGlance is in Fewer or More Details Mode. Regardless, go to the Tools tab and click Protein Crosslinks:
 |
3. At the list of crosslink types in the current molecule, click on Isopeptides:
 |
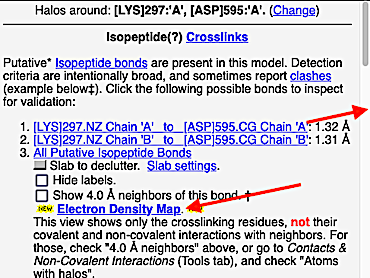
4. At the list of putative crosslink bonds, click on the first case.
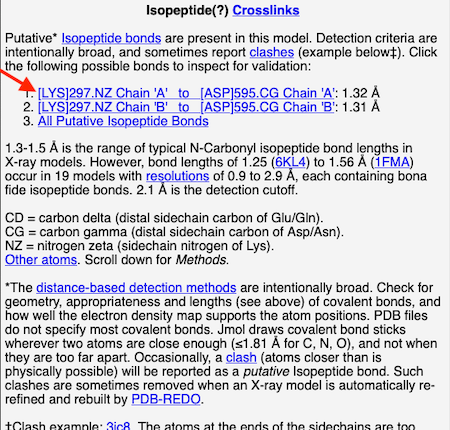 |
5. A brief movie will play, showing you where in the entire structure this particular pair of residues is positioned, then zooming in to show you the pair in detail. As FirstGlance explains, your job is to decide whether this is a genuine isopeptide bond, or not (a clash-error in the model). This pair of residues is modeled as an isopeptide bond (not as two unconnected residues clashing). The isopeptide bond length of 1.3 Å is appropriate for an isopeptide bond (see paragraph about bond lengths snapshot at step #4). The Abstract of the publication (click on "Abstract at PubMed" near the bottom of the Molecule Information Tab) mentions both the thioester and isopeptide bonds detected by FirstGlance.
|
C |
6. Finally, click on the link to show the Electron Density Map. At the very good resolution of 1.9 Å, every atom in this pair of residues has clear electron density. There is no ambiguity about where these atoms belong in the model. Therefore, the electron density map supports the presence of this isopeptide bond.
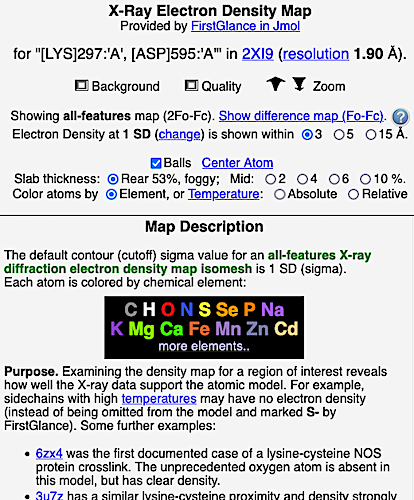 | 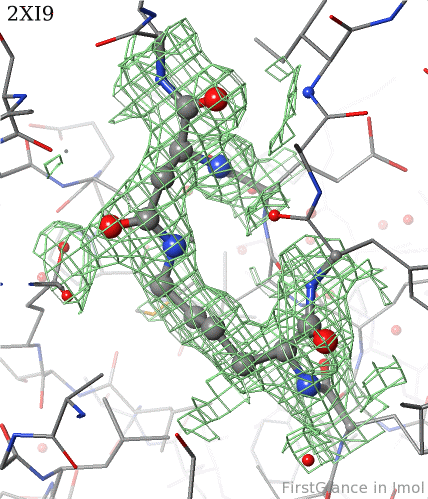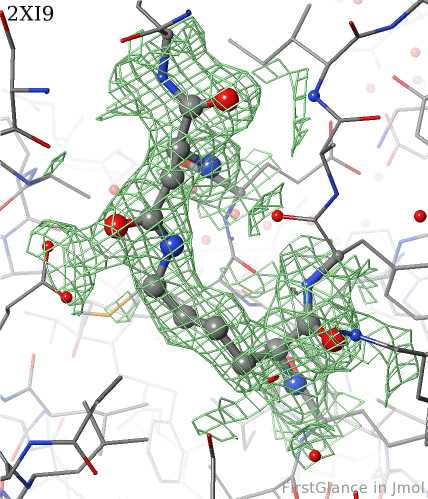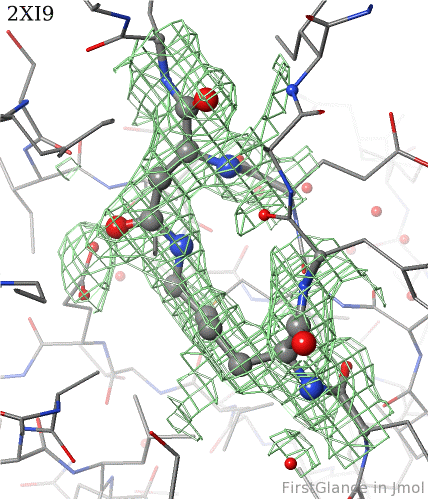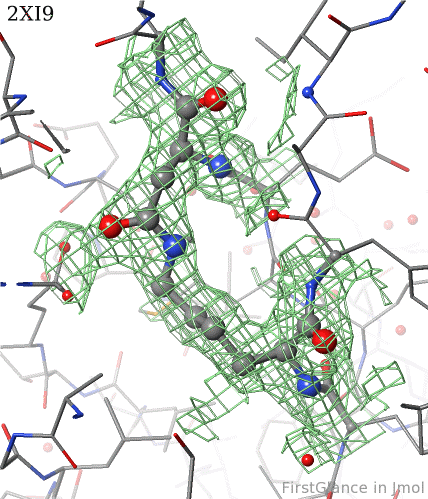 |
Conservation of Crosslinking ResiduesConservation of Crosslinking Residues
If the PDB file is pre-processed by ConSurf, after displaying the crosslinked residues, their levels of evolutionary conservation will be shown by checking ConSurf Colors (in the middle-left of FirstGlance).
For more complete instructions and illustrations, see Interpreting ConSurf Results.
For example, below is a thioether covalent crosslink between Cys356 and Tyr379 in catalase 1sy7. (The multiple sequence alignment included 150 sequences sampled from 22,855 unique similar sequences. The average pairwise distance was 0.96.)
|
Thioether crosslink in catalase 1sy7.
|
Crosslinking residues:
|
Additional crosslink examples showing conservation:
Verified Crosslink ExamplesVerified Crosslink Examples
Examples of verified crosslinks of all 7 types detected by FirstGlance are listed within FirstGlance. Display the list of crosslink types by clicking Protein Crosslinks in the Tools tab (as shown in step #2 above). Regardless of whether the model being displayed has any crosslinks, clicking on each type of crosslink in turn will display further information, including examples, in the lower left panel. For example, here is a snapshot of the examples listed by FirstGlance for Histidine-Tyrosine crosslinks:
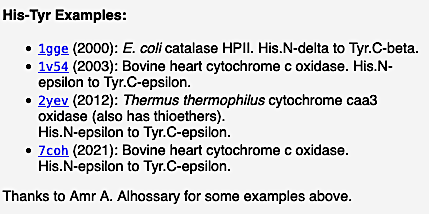 |
ClashesClashes
FirstGlance suggests the presence of a "putative crosslink" based solely on the distances between relevant atoms (click the link Detailed Methods in FirstGlance). Sometimes, instead of a real covalent bond crosslink, the proximity of the relevant atoms is due to alternate locations or small errors in the atomic model that produce clashes.
For each putative crosslink suggested by FirstGlance, it is the responsibility of the user to evaluate whether it is a real covalent crosslink, or a clash. FirstGlance makes this responsibility clear (see the bottom paragraph in the snapshot under step #4 above).
BondsBonds
It is important not to take the bonds displayed by FirstGlance/Jmol literally. Atomic coordinate files from the wwPDB do not specify most covalent bonds. Rather, Jmol draws covalent "bond" sticks between atoms wherever two atoms are sufficiently close. In the case of carbon, nitrogen and oxygen, this is wherever a pair of those atoms is separated by ≤1.81 Å (for actual bond lengths, see footnote[1]). In perfectly accurate models, this works correctly, but the errors in empirical models sometimes lead to bonds being drawn incorrectly. Thus, Jmol may show bond sticks where the reality is a clash, not a bond; and Jmol may fail to show bond sticks between atoms that are believed to be covalently bonded, but are too far apart in the model.
Case IllustrationsCase Illustrations
For each type of crosslink, FirstGlance gives one or more examples of clashes just above the list of verified crosslink examples, explaining the evidence for a clash. Below are a few specific cases.
His-TyrHis-Tyr
His-Tyr Genuine CrosslinkHis-Tyr Genuine Crosslink
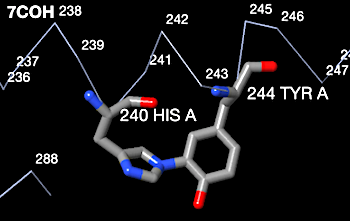
Histidine-tyrosine covalent crosslinks are known to occur in some c-type cytochromes. Genuine His-Tyr crosslinks occur, for example, in bovine heart cytochrome C oxidase 7coh. The snapshot below from FirstGlance shows the correct geometry for a genuine His-Tyr covalent crosslink. The His nitrogen is 1.5 Å from the Tyr carbon. This compares well with the typical C-N covalent bond length of 1.4 Å[1].
|
C |
His-Tyr ClashHis-Tyr Clash
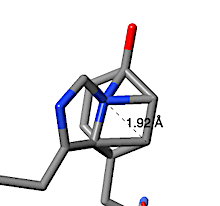
In contrast, the putative His-Tyr crosslink suggested by FirstGlance to be in human nuclease 4rea turns out to be a clash. The text-snapshot below at right is the explanation from FirstGlance. When a His sidechain nitrogen is within 2.2 Å of a Tyr sidechain carbon, FirstGlance suggests a putative His-Tyr crosslink. The cutoff distance is generous to accommodate some errors in the model, but also increases the chances of reporting a clash. In the case of 4rea, a His sidechain nitrogen is 1.92 Å from a Tyr sidechain carbon as a result of these two sidechains being positioned impossibly close to each other.
|
C |
4rea also has a clash suggested as a possible isopeptide bond by FirstGlance. Both that, and the His-Tyr clashes are removed by PDB REDO.
ThioestersThioesters
Thioester Genuine CrosslinkThioester Genuine Crosslink
|
C |
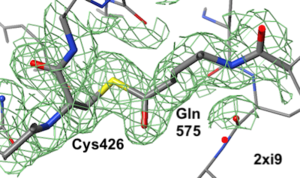
2xi9 has 2 copies of a bacterial pilus adhesin in its asymmetric unit. FirstGlance reports a putative thioester crosslink in each copy of the protein. The relevant residues are modeled as a thiester bond with correct geometry. The C-S distances in the model are 1.6 Å, in good agreement with the typical bond length of 1.7 Å[1]. The electron density map shows clear density for the thioester-bonded atoms. The difference map (not shown) raises no alarms. The abstract of the publication highlights this thioester bond.
"Thioester" Clash"Thioester" Clash
|
Electron density maps
|
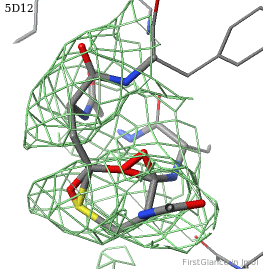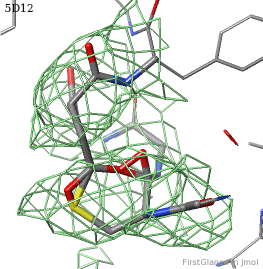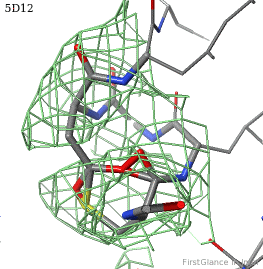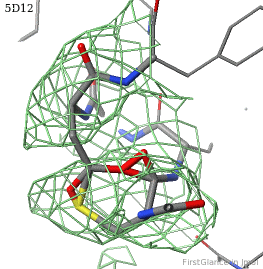
5d12 has two copies of a kinase domain in its asymmetric unit. FirstGlance reports a possible thioester crosslink only in chain B. The C-S distance is 1.76 Å, quite reasonable. However, the residues are not modeled as a thiester bond, and Jmol draws many spurious "bonds". The sidechain of Asp348 passes through a gap in the all-features electron density (green isomesh, above left). The difference electron density map (above right) reveals insufficient density to account for the position of the Asp sidechain (dark red isomesh), while suggesting an alternative position for those sidechain atoms (blue isomesh). In conclusion, this is a clash, not a thioester crosslink bond.
After PDB REDO re-refines and rebuilds 5d12, this clash is absent.
Lys-Cys NOSLys-Cys NOS
Lys-Cys NOS Genuine CrosslinksLys-Cys NOS Genuine Crosslinks
The Lys-Cys NOS protein crosslink bond was first suggested in 2016, and firmly established in 2021 (see Lysine-cysteine NOS bonds). NOS crosslinks are allosteric redox switches[2]. The first crystal structure with an NOS bond that models all 3 of the bonded NOS atoms is 6zx4 (2021; electron density map shown at Electron_density_maps#2Fo-Fc_Map).
Since NOS bonds were not well recognized before 2021, earlier crystal structures containing them typically omitted the oxygen. An example is 3u7z. See the electron density map shown at Electron_density_maps#2Fo-Fc_Map, bearing in mind that the 2Fo-Fc density for an atom missing in the model will be half of the density for an atom present in the model. The electron density difference map (Fo-Fc) clearly indicates that an atom is missing between the lysine sidechain nitrogen and the cysteine sulfur (Electron_density_maps#Fo-Fc_Difference_Map).
Lys-Cys Non-CrosslinksLys-Cys Non-Crosslinks
There are cases where the sidechain nitrogen of a Lysine is the right distance from the sulfur of a cysteine to raise the possibility of an NOS bond, but there is no such bond. When the distance is ≤ 3.1 Å, FirstGlance suggests the possiblility. If the electron density map is clear in the area of the possible bond, its presence or absence can be determined. 6vb2 has lysine sidechain nitrogens 2.6-2.7 Å from cysteine sulfurs, but the electron density map (Electron_density_maps#2Fo-Fc_Map) shows that there is no oxygen between these atoms.
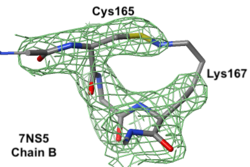
Lys-Cys ClashLys-Cys Clash
 |
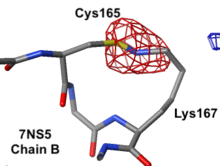 |
Electron density maps
|
7ns5 is a homo-tetramer. FirstGlance suggests a possible Lys-Cys NOS crosslink in chain B. The fact that FirstGlance suggests no such crosslinks in the other 3 chains is a hint that the proximity of the sidechain nitrogen of Lys167 to the sulfur in Cys165 in chain B is a clash. The atomic model shows an improbable N-S bond between the sidechains. Further, electron density (above left) is missing for the last 3 atoms of the lysine sidechain, consistent with it being misplaced (green isomesh). The difference electron density map (above right) reveals that there is no electron density consistent with the lysine sidechain nitrogen being located where it was placed in the model (dark red isomesh). In conclusion, this is a clash in chain B, not an NOS crosslink.
After PDB REDO re-refines and rebuilds 7ns5, this clash is absent.
See AlsoSee Also
- FirstGlance/Index: FirstGlance resources within and external to Proteopedia.
Notes and ReferencesNotes and References
- ↑ 1.0 1.1 1.2 Actual covalent bond lengths are approximately C-C: 1.4 Å; C-N: 1.4 Å; C-O: 1.4 Å; C=O: 1.2 Å; C-S: 1.7 Å (Covalent radii in Wikipedia).
- ↑ Wensien M, von Pappenheim FR, Funk LM, Kloskowski P, Curth U, Diederichsen U, Uranga J, Ye J, Fang P, Pan KT, Urlaub H, Mata RA, Sautner V, Tittmann K. A lysine-cysteine redox switch with an NOS bridge regulates enzyme function. Nature. 2021 May 5. pii: 10.1038/s41586-021-03513-3. doi:, 10.1038/s41586-021-03513-3. PMID:33953398 doi:http://dx.doi.org/10.1038/s41586-021-03513-3