2oui: Difference between revisions
m Protected "2oui" [edit=sysop:move=sysop] |
No edit summary |
||
| (10 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
< | ==D275P mutant of alcohol dehydrogenase from protozoa Entamoeba histolytica== | ||
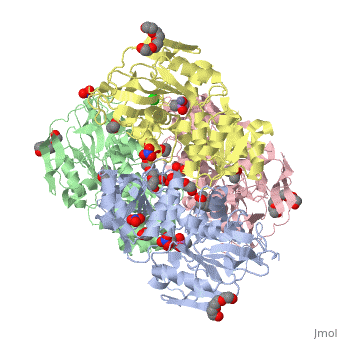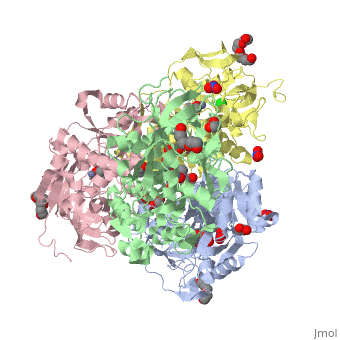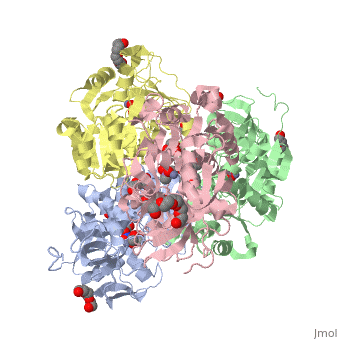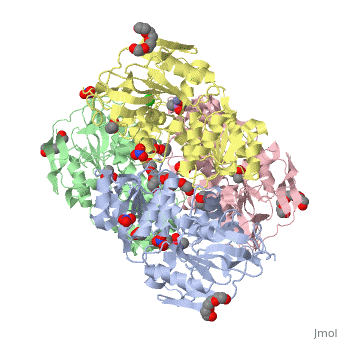
<StructureSection load='2oui' size='340' side='right'caption='[[2oui]], [[Resolution|resolution]] 1.77Å' scene=''> | |||
You may | == Structural highlights == | ||
<table><tr><td colspan='2'>[[2oui]] is a 4 chain structure with sequence from [https://en.wikipedia.org/wiki/Entamoeba_histolytica Entamoeba histolytica]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=2OUI OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=2OUI FirstGlance]. <br> | |||
</td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 1.77Å</td></tr> | |||
-- | <tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=1PE:PENTAETHYLENE+GLYCOL'>1PE</scene>, <scene name='pdbligand=CAC:CACODYLATE+ION'>CAC</scene>, <scene name='pdbligand=CL:CHLORIDE+ION'>CL</scene>, <scene name='pdbligand=EDO:1,2-ETHANEDIOL'>EDO</scene>, <scene name='pdbligand=NO3:NITRATE+ION'>NO3</scene>, <scene name='pdbligand=PG4:TETRAETHYLENE+GLYCOL'>PG4</scene>, <scene name='pdbligand=PGE:TRIETHYLENE+GLYCOL'>PGE</scene>, <scene name='pdbligand=ZN:ZINC+ION'>ZN</scene></td></tr> | ||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=2oui FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=2oui OCA], [https://pdbe.org/2oui PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=2oui RCSB], [https://www.ebi.ac.uk/pdbsum/2oui PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=2oui ProSAT]</span></td></tr> | |||
</table> | |||
== Function == | |||
[https://www.uniprot.org/uniprot/ADH1_ENTHI ADH1_ENTHI] Alcohol dehydrogenase with a preference for medium chain secondary alcohols, such as 2-butanol and isopropanol. Has very low activity with primary alcohols, such as ethanol. Under physiological conditions, the enzyme reduces aldehydes and 2-ketones to produce secondary alcohols. Is also active with acetaldehyde and propionaldehyde.<ref>PMID:20102159</ref> | |||
== Evolutionary Conservation == | |||
[[Image:Consurf_key_small.gif|200px|right]] | |||
Check<jmol> | |||
<jmolCheckbox> | |||
<scriptWhenChecked>; select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/ou/2oui_consurf.spt"</scriptWhenChecked> | |||
<scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> | |||
<text>to colour the structure by Evolutionary Conservation</text> | |||
</jmolCheckbox> | |||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=2oui ConSurf]. | |||
<div style="clear:both"></div> | |||
<div style="background-color:#fffaf0;"> | |||
== Publication Abstract from PubMed == | |||
Analysis of the three-dimensional structures of two closely related thermophilic and hyperthermophilic alcohol dehydrogenases (ADHs) from the respective microorganisms Entamoeba histolytica (EhADH1) and Thermoanaerobacter brockii (TbADH) suggested that a unique, strategically located proline residue (Pro275) at the center of the dimerization interface might be crucial for maintaining the thermal stability of TbADH. To assess the contribution of Pro275 to the thermal stability of the ADHs, we applied site-directed mutagenesis to replace Asp275 of EhADH1 with Pro (D275P-EhADH1) and conversely Pro275 of TbADH with Asp (P275D-TbADH). The results indicate that replacing Asp275 with Pro significantly enhances the thermal stability of EhADH1 (DeltaT(1/2) </= +10 degrees C), whereas the reverse mutation in the thermophilic TbADH (P275D-TbADH) reduces the thermostability of the enzyme (DeltaT(1/2) </= -18.8 degrees C). Analysis of the crystal structures of the thermostabilized mutant D275P-EhADH1 and the thermocompromised mutant P275D-TbADH suggest that a proline residue at position 275 thermostabilized the enzymes by reducing flexibility and by reinforcing hydrophobic interactions at the dimer-dimer interface of the tetrameric ADHs. Proteins 2008. (c) 2008 Wiley-Liss, Inc. | |||
Thermal stabilization of the protozoan Entamoeba histolytica alcohol dehydrogenase by a single proline substitution.,Goihberg E, Dym O, Tel-Or S, Shimon L, Frolow F, Peretz M, Burstein Y Proteins. 2008 Feb 7;. PMID:18260103<ref>PMID:18260103</ref> | |||
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | |||
</div> | |||
<div class="pdbe-citations 2oui" style="background-color:#fffaf0;"></div> | |||
== | |||
==See Also== | ==See Also== | ||
*[[Alcohol dehydrogenase|Alcohol dehydrogenase]] | *[[Alcohol dehydrogenase|Alcohol dehydrogenase]] | ||
*[[ | *[[Alcohol dehydrogenase 3D structures|Alcohol dehydrogenase 3D structures]] | ||
== References == | |||
== | <references/> | ||
< | __TOC__ | ||
</StructureSection> | |||
[[Category: Entamoeba histolytica]] | [[Category: Entamoeba histolytica]] | ||
[[Category: Burstein | [[Category: Large Structures]] | ||
[[Category: Dym | [[Category: Burstein Y]] | ||
[[Category: Frolow | [[Category: Dym O]] | ||
[[Category: Goihberg | [[Category: Frolow F]] | ||
[[Category: Peretz | [[Category: Goihberg E]] | ||
[[Category: Shimon | [[Category: Peretz M]] | ||
[[Category: Shimon L]] | |||
Latest revision as of 11:58, 25 October 2023
D275P mutant of alcohol dehydrogenase from protozoa Entamoeba histolyticaD275P mutant of alcohol dehydrogenase from protozoa Entamoeba histolytica
Structural highlights
FunctionADH1_ENTHI Alcohol dehydrogenase with a preference for medium chain secondary alcohols, such as 2-butanol and isopropanol. Has very low activity with primary alcohols, such as ethanol. Under physiological conditions, the enzyme reduces aldehydes and 2-ketones to produce secondary alcohols. Is also active with acetaldehyde and propionaldehyde.[1] Evolutionary Conservation Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. Publication Abstract from PubMedAnalysis of the three-dimensional structures of two closely related thermophilic and hyperthermophilic alcohol dehydrogenases (ADHs) from the respective microorganisms Entamoeba histolytica (EhADH1) and Thermoanaerobacter brockii (TbADH) suggested that a unique, strategically located proline residue (Pro275) at the center of the dimerization interface might be crucial for maintaining the thermal stability of TbADH. To assess the contribution of Pro275 to the thermal stability of the ADHs, we applied site-directed mutagenesis to replace Asp275 of EhADH1 with Pro (D275P-EhADH1) and conversely Pro275 of TbADH with Asp (P275D-TbADH). The results indicate that replacing Asp275 with Pro significantly enhances the thermal stability of EhADH1 (DeltaT(1/2) </= +10 degrees C), whereas the reverse mutation in the thermophilic TbADH (P275D-TbADH) reduces the thermostability of the enzyme (DeltaT(1/2) </= -18.8 degrees C). Analysis of the crystal structures of the thermostabilized mutant D275P-EhADH1 and the thermocompromised mutant P275D-TbADH suggest that a proline residue at position 275 thermostabilized the enzymes by reducing flexibility and by reinforcing hydrophobic interactions at the dimer-dimer interface of the tetrameric ADHs. Proteins 2008. (c) 2008 Wiley-Liss, Inc. Thermal stabilization of the protozoan Entamoeba histolytica alcohol dehydrogenase by a single proline substitution.,Goihberg E, Dym O, Tel-Or S, Shimon L, Frolow F, Peretz M, Burstein Y Proteins. 2008 Feb 7;. PMID:18260103[2] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences
|
| ||||||||||||||||||