5cgn: Difference between revisions
No edit summary |
No edit summary |
||
| (2 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
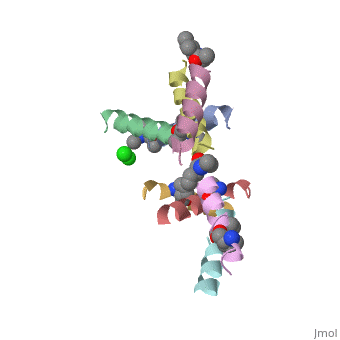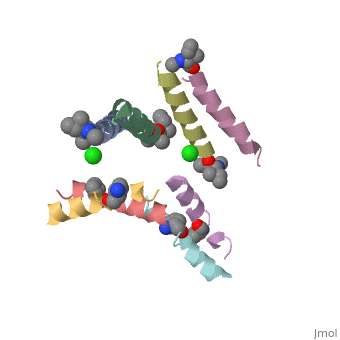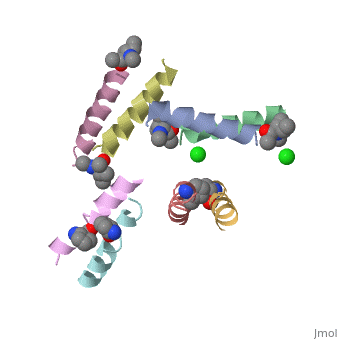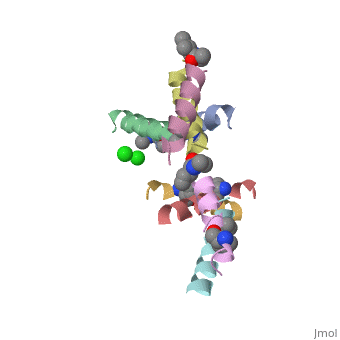
==Structure of quasiracemic Ala-Magainin 2 with a beta amino acid substitution at position 8== | ==Structure of quasiracemic Ala-Magainin 2 with a beta amino acid substitution at position 8== | ||
<StructureSection load='5cgn' size='340' side='right' caption='[[5cgn]], [[Resolution|resolution]] 2.20Å' scene=''> | <StructureSection load='5cgn' size='340' side='right'caption='[[5cgn]], [[Resolution|resolution]] 2.20Å' scene=''> | ||
== Structural highlights == | == Structural highlights == | ||
<table><tr><td colspan='2'>[[5cgn]] is a 8 chain structure. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=5CGN OCA]. For a <b>guided tour on the structure components</b> use [ | <table><tr><td colspan='2'>[[5cgn]] is a 8 chain structure with sequence from [https://en.wikipedia.org/wiki/Synthetic_construct Synthetic construct]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=5CGN OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=5CGN FirstGlance]. <br> | ||
</td></tr><tr id=' | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 2.2Å</td></tr> | ||
<tr id=' | <tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=CL:CHLORIDE+ION'>CL</scene>, <scene name='pdbligand=DAL:D-ALANINE'>DAL</scene>, <scene name='pdbligand=DGL:D-GLUTAMIC+ACID'>DGL</scene>, <scene name='pdbligand=DHI:D-HISTIDINE'>DHI</scene>, <scene name='pdbligand=DIL:D-ISOLEUCINE'>DIL</scene>, <scene name='pdbligand=DLE:D-LEUCINE'>DLE</scene>, <scene name='pdbligand=DLY:D-LYSINE'>DLY</scene>, <scene name='pdbligand=DPN:D-PHENYLALANINE'>DPN</scene>, <scene name='pdbligand=DSE:N-METHYL-D-SERINE'>DSE</scene>, <scene name='pdbligand=DSG:D-ASPARAGINE'>DSG</scene>, <scene name='pdbligand=DVA:D-VALINE'>DVA</scene>, <scene name='pdbligand=MED:D-METHIONINE'>MED</scene>, <scene name='pdbligand=XCP:(1S,2S)-2-AMINOCYCLOPENTANECARBOXYLIC+ACID'>XCP</scene></td></tr> | ||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=5cgn FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=5cgn OCA], [https://pdbe.org/5cgn PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=5cgn RCSB], [https://www.ebi.ac.uk/pdbsum/5cgn PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=5cgn ProSAT]</span></td></tr> | |||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[ | |||
</table> | </table> | ||
== Function == | |||
[https://www.uniprot.org/uniprot/MAGA_XENLA MAGA_XENLA] Antimicrobial peptides that inhibit the growth of numerous species of bacteria and fungi and induce osmotic lysis of protozoa. Magainins are membrane lytic agents. | |||
<div style="background-color:#fffaf0;"> | <div style="background-color:#fffaf0;"> | ||
== Publication Abstract from PubMed == | == Publication Abstract from PubMed == | ||
| Line 18: | Line 19: | ||
</div> | </div> | ||
<div class="pdbe-citations 5cgn" style="background-color:#fffaf0;"></div> | <div class="pdbe-citations 5cgn" style="background-color:#fffaf0;"></div> | ||
==See Also== | |||
*[[Magainin 2|Magainin 2]] | |||
== References == | == References == | ||
<references/> | <references/> | ||
__TOC__ | __TOC__ | ||
</StructureSection> | </StructureSection> | ||
[[Category: | [[Category: Large Structures]] | ||
[[Category: | [[Category: Synthetic construct]] | ||
[[Category: | [[Category: Forest KT]] | ||
[[Category: | [[Category: Gellman SH]] | ||
[[Category: | [[Category: Hayouka Z]] | ||
[[Category: | [[Category: Mortenson DE]] | ||
[[Category: | [[Category: Satyshur KA]] | ||
[[Category: | [[Category: Thomas NC]] | ||
[[Category: | [[Category: Weisblum B]] | ||
Latest revision as of 11:37, 27 September 2023
Structure of quasiracemic Ala-Magainin 2 with a beta amino acid substitution at position 8Structure of quasiracemic Ala-Magainin 2 with a beta amino acid substitution at position 8
Structural highlights
FunctionMAGA_XENLA Antimicrobial peptides that inhibit the growth of numerous species of bacteria and fungi and induce osmotic lysis of protozoa. Magainins are membrane lytic agents. Publication Abstract from PubMedQuasiracemic crystallography has been used to explore the significance of homochiral and heterochiral associations in a set of host-defense peptide derivatives. The previously reported racemic crystal structure of a magainin 2 derivative displayed a homochiral antiparallel dimer association featuring a "phenylalanine zipper" notable for the dual roles of phenylalanines in mediating dimerization and formation of an exposed hydrophobic swath. This motif is seen as well in two new quasiracemate crystals that contain the d form of the magainin 2 derivative along with an l-peptide in which one Ala has been replaced by a beta-amino acid residue. This structural trend supports the hypothesis that the Phe zipper motif has functional significance. Quasiracemate Crystal Structures of Magainin 2 Derivatives Support the Functional Significance of the Phenylalanine Zipper Motif.,Hayouka Z, Thomas NC, Mortenson DE, Satyshur KA, Weisblum B, Forest KT, Gellman SH J Am Chem Soc. 2015 Sep 23;137(37):11884-7. doi: 10.1021/jacs.5b07206. Epub 2015 , Sep 10. PMID:26369301[1] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences
|
| ||||||||||||||||||