1s5o: Difference between revisions
No edit summary |
No edit summary |
||
| (13 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
==Structural and Mutational Characterization of L-carnitine Binding to Human carnitine Acetyltransferase== | |||
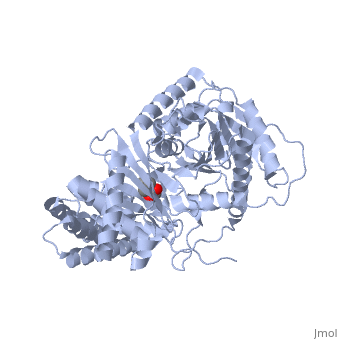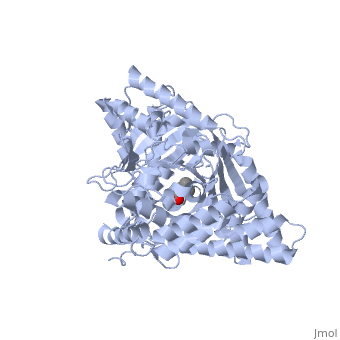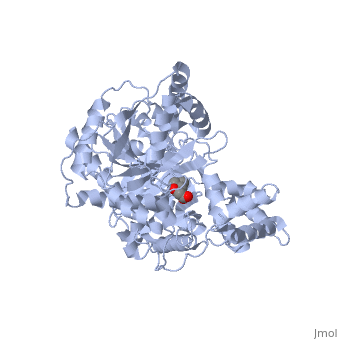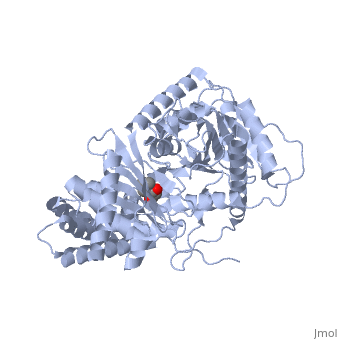
<StructureSection load='1s5o' size='340' side='right'caption='[[1s5o]], [[Resolution|resolution]] 1.80Å' scene=''> | |||
== Structural highlights == | |||
<table><tr><td colspan='2'>[[1s5o]] is a 1 chain structure with sequence from [https://en.wikipedia.org/wiki/Homo_sapiens Homo sapiens]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1S5O OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=1S5O FirstGlance]. <br> | |||
</td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 1.8Å</td></tr> | |||
| | <tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=152:CARNITINE'>152</scene></td></tr> | ||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1s5o FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1s5o OCA], [https://pdbe.org/1s5o PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1s5o RCSB], [https://www.ebi.ac.uk/pdbsum/1s5o PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1s5o ProSAT]</span></td></tr> | |||
</table> | |||
== Function == | |||
[https://www.uniprot.org/uniprot/CACP_HUMAN CACP_HUMAN] Carnitine acetylase is specific for short chain fatty acids. Carnitine acetylase seems to affect the flux through the pyruvate dehydrogenase complex. It may be involved as well in the transport of acetyl-CoA into mitochondria. | |||
== Evolutionary Conservation == | |||
[[Image:Consurf_key_small.gif|200px|right]] | |||
Check<jmol> | |||
<jmolCheckbox> | |||
<scriptWhenChecked>; select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/s5/1s5o_consurf.spt"</scriptWhenChecked> | |||
<scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> | |||
<text>to colour the structure by Evolutionary Conservation</text> | |||
</jmolCheckbox> | |||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1s5o ConSurf]. | |||
<div style="clear:both"></div> | |||
<div style="background-color:#fffaf0;"> | |||
== Publication Abstract from PubMed == | |||
We report the crystal structure of a binary complex of human peroxisomal carnitine acetyltransferase and the substrate l-carnitine, refined to a resolution of 1.8 Angstrom with an R(factor) value of 18.9% (R(free)=22.3%). L-carnitine binds to a preformed pocket in the active site tunnel of carnitine acetyltransferase aligned with His(322). The quaternary nitrogen of carnitine forms a pi-cation interaction with Phe(545), while Arg(497) forms an electrostatic interaction with the negatively charged carboxylate group. An extensive hydrogen bond network also occurs between the carboxylate group and Tyr(431), Thr(444), and a bound water molecule. Site-directed mutagenesis and kinetic characterization reveals that Tyr(431), Thr(444), Arg(497), and Phe(545) are essential for high affinity binding of L-carnitine. | |||
Structural and mutational characterization of L-carnitine binding to human carnitine acetyltransferase.,Govindasamy L, Kukar T, Lian W, Pedersen B, Gu Y, Agbandje-McKenna M, Jin S, McKenna R, Wu D J Struct Biol. 2004 Jun;146(3):416-24. PMID:15099582<ref>PMID:15099582</ref> | |||
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | |||
</div> | |||
<div class="pdbe-citations 1s5o" style="background-color:#fffaf0;"></div> | |||
== | ==See Also== | ||
*[[Carnitine acetyltransferase|Carnitine acetyltransferase]] | |||
== References == | |||
== | <references/> | ||
__TOC__ | |||
</StructureSection> | |||
[[Category: Homo sapiens]] | [[Category: Homo sapiens]] | ||
[[Category: | [[Category: Large Structures]] | ||
[[Category: Agbandje-Mckenna | [[Category: Agbandje-Mckenna M]] | ||
[[Category: Govindasamy | [[Category: Govindasamy L]] | ||
[[Category: Gu | [[Category: Gu Y]] | ||
[[Category: Jin | [[Category: Jin S]] | ||
[[Category: Kukar | [[Category: Kukar T]] | ||
[[Category: Lian | [[Category: Lian W]] | ||
[[Category: Mckenna | [[Category: Mckenna R]] | ||
[[Category: Pedersen | [[Category: Pedersen B]] | ||
[[Category: Wu | [[Category: Wu D]] | ||
Latest revision as of 09:13, 23 August 2023
Structural and Mutational Characterization of L-carnitine Binding to Human carnitine AcetyltransferaseStructural and Mutational Characterization of L-carnitine Binding to Human carnitine Acetyltransferase
Structural highlights
FunctionCACP_HUMAN Carnitine acetylase is specific for short chain fatty acids. Carnitine acetylase seems to affect the flux through the pyruvate dehydrogenase complex. It may be involved as well in the transport of acetyl-CoA into mitochondria. Evolutionary Conservation Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. Publication Abstract from PubMedWe report the crystal structure of a binary complex of human peroxisomal carnitine acetyltransferase and the substrate l-carnitine, refined to a resolution of 1.8 Angstrom with an R(factor) value of 18.9% (R(free)=22.3%). L-carnitine binds to a preformed pocket in the active site tunnel of carnitine acetyltransferase aligned with His(322). The quaternary nitrogen of carnitine forms a pi-cation interaction with Phe(545), while Arg(497) forms an electrostatic interaction with the negatively charged carboxylate group. An extensive hydrogen bond network also occurs between the carboxylate group and Tyr(431), Thr(444), and a bound water molecule. Site-directed mutagenesis and kinetic characterization reveals that Tyr(431), Thr(444), Arg(497), and Phe(545) are essential for high affinity binding of L-carnitine. Structural and mutational characterization of L-carnitine binding to human carnitine acetyltransferase.,Govindasamy L, Kukar T, Lian W, Pedersen B, Gu Y, Agbandje-McKenna M, Jin S, McKenna R, Wu D J Struct Biol. 2004 Jun;146(3):416-24. PMID:15099582[1] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences
|
| ||||||||||||||||||