1puf: Difference between revisions
No edit summary |
No edit summary |
||
| (4 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
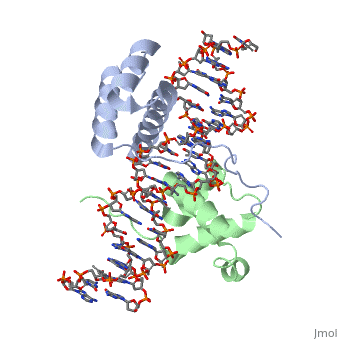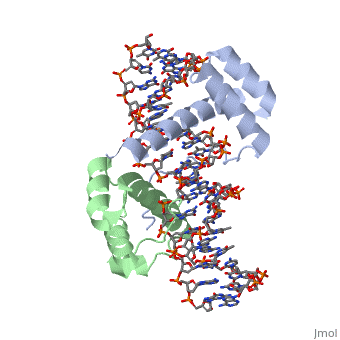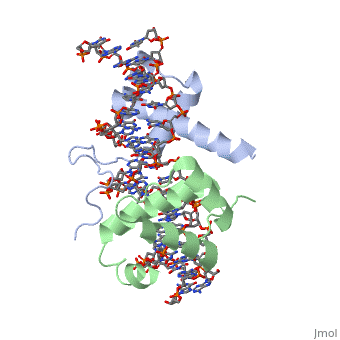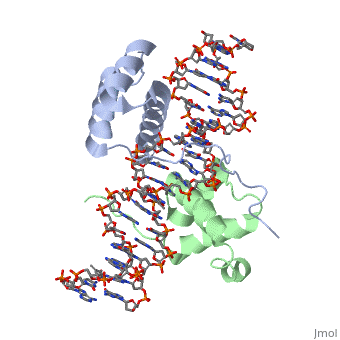
==Crystal Structure of HoxA9 and Pbx1 homeodomains bound to DNA== | ==Crystal Structure of HoxA9 and Pbx1 homeodomains bound to DNA== | ||
<StructureSection load='1puf' size='340' side='right' caption='[[1puf]], [[Resolution|resolution]] 1.90Å' scene=''> | <StructureSection load='1puf' size='340' side='right'caption='[[1puf]], [[Resolution|resolution]] 1.90Å' scene=''> | ||
== Structural highlights == | == Structural highlights == | ||
<table><tr><td colspan='2'>[[1puf]] is a 4 chain structure with sequence from [ | <table><tr><td colspan='2'>[[1puf]] is a 4 chain structure with sequence from [https://en.wikipedia.org/wiki/Homo_sapiens Homo sapiens] and [https://en.wikipedia.org/wiki/Mus_musculus Mus musculus]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1PUF OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=1PUF FirstGlance]. <br> | ||
</td></tr><tr><td class="sblockLbl"><b>[[ | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 1.9Å</td></tr> | ||
<tr><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[ | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1puf FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1puf OCA], [https://pdbe.org/1puf PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1puf RCSB], [https://www.ebi.ac.uk/pdbsum/1puf PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1puf ProSAT]</span></td></tr> | ||
<table> | </table> | ||
== Function == | == Function == | ||
[ | [https://www.uniprot.org/uniprot/HXA9_MOUSE HXA9_MOUSE] Sequence-specific transcription factor which is part of a developmental regulatory system that provides cells with specific positional identities on the anterior-posterior axis. Required for induction of E-selectin and VCAM-1, on the endothelial cells surface at sites of inflammation (By similarity). | ||
== Evolutionary Conservation == | == Evolutionary Conservation == | ||
[[Image:Consurf_key_small.gif|200px|right]] | [[Image:Consurf_key_small.gif|200px|right]] | ||
Check<jmol> | Check<jmol> | ||
<jmolCheckbox> | <jmolCheckbox> | ||
<scriptWhenChecked>select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/pu/1puf_consurf.spt"</scriptWhenChecked> | <scriptWhenChecked>; select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/pu/1puf_consurf.spt"</scriptWhenChecked> | ||
<scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> | <scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> | ||
<text>to colour the structure by Evolutionary Conservation</text> | <text>to colour the structure by Evolutionary Conservation</text> | ||
</jmolCheckbox> | </jmolCheckbox> | ||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/ | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1puf ConSurf]. | ||
<div style="clear:both"></div> | <div style="clear:both"></div> | ||
<div style="background-color:#fffaf0;"> | <div style="background-color:#fffaf0;"> | ||
| Line 28: | Line 27: | ||
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | ||
</div> | </div> | ||
<div class="pdbe-citations 1puf" style="background-color:#fffaf0;"></div> | |||
==See Also== | ==See Also== | ||
*[[Hox protein|Hox protein]] | |||
*[[Tomato aspermy virus protein 2b Suppression of RNA Silencing|Tomato aspermy virus protein 2b Suppression of RNA Silencing]] | *[[Tomato aspermy virus protein 2b Suppression of RNA Silencing|Tomato aspermy virus protein 2b Suppression of RNA Silencing]] | ||
== References == | == References == | ||
<references/> | <references/> | ||
| Line 37: | Line 37: | ||
</StructureSection> | </StructureSection> | ||
[[Category: Homo sapiens]] | [[Category: Homo sapiens]] | ||
[[Category: Large Structures]] | |||
[[Category: Mus musculus]] | [[Category: Mus musculus]] | ||
[[Category: Laronde-Leblanc | [[Category: Laronde-Leblanc NA]] | ||
[[Category: Wolberger | [[Category: Wolberger C]] | ||
Latest revision as of 12:48, 16 August 2023
Crystal Structure of HoxA9 and Pbx1 homeodomains bound to DNACrystal Structure of HoxA9 and Pbx1 homeodomains bound to DNA
Structural highlights
FunctionHXA9_MOUSE Sequence-specific transcription factor which is part of a developmental regulatory system that provides cells with specific positional identities on the anterior-posterior axis. Required for induction of E-selectin and VCAM-1, on the endothelial cells surface at sites of inflammation (By similarity). Evolutionary Conservation Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. Publication Abstract from PubMedThe HOX/HOM superfamily of homeodomain proteins controls cell fate and segmental embryonic patterning by a mechanism that is conserved in all metazoans. The linear arrangement of the Hox genes on the chromosome correlates with the spatial distribution of HOX protein expression along the anterior-posterior axis of the embryo. Most HOX proteins bind DNA cooperatively with members of the PBC family of TALE-type homeodomain proteins, which includes human Pbx1. Cooperative DNA binding between HOX and PBC proteins requires a residue N-terminal to the HOX homeodomain termed the hexapeptide, which differs significantly in sequence between anterior- and posterior-regulating HOX proteins. We report here the 1.9-A-resolution structure of a posterior HOX protein, HoxA9, complexed with Pbx1 and DNA, which reveals that the posterior Hox hexapeptide adopts an altered conformation as compared with that seen in previously determined anterior HOX/PBC structures. The additional nonspecific interactions and altered DNA conformation in this structure account for the stronger DNA-binding affinity and altered specificity observed for posterior HOX proteins when compared with anterior HOX proteins. DNA-binding studies of wild-type and mutant HoxA9 and HoxB1 show residues in the N-terminal arm of the homeodomains are critical for proper DNA sequence recognition despite lack of direct contact by these residues to the DNA bases. These results help shed light on the mechanism of transcriptional regulation by HOX proteins and show how DNA-binding proteins may use indirect contacts to determine sequence specificity. Structure of HoxA9 and Pbx1 bound to DNA: Hox hexapeptide and DNA recognition anterior to posterior.,LaRonde-LeBlanc NA, Wolberger C Genes Dev. 2003 Aug 15;17(16):2060-72. PMID:12923056[1] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences
|
| ||||||||||||||||