1b41: Difference between revisions
No edit summary |
No edit summary |
||
| (10 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
< | ==HUMAN ACETYLCHOLINESTERASE COMPLEXED WITH FASCICULIN-II, GLYCOSYLATED PROTEIN== | ||
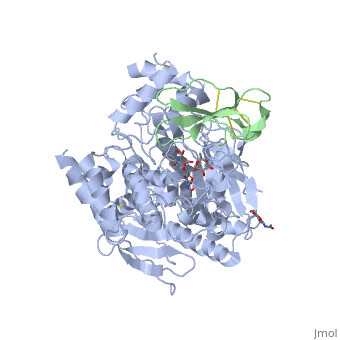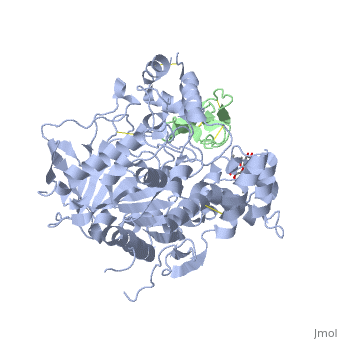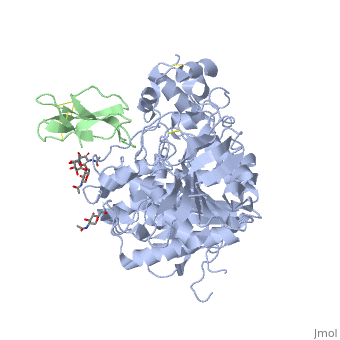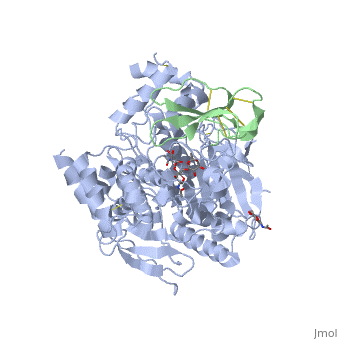
<StructureSection load='1b41' size='340' side='right'caption='[[1b41]], [[Resolution|resolution]] 2.76Å' scene=''> | |||
You may | == Structural highlights == | ||
<table><tr><td colspan='2'>[[1b41]] is a 2 chain structure with sequence from [https://en.wikipedia.org/wiki/Dendroaspis_angusticeps Dendroaspis angusticeps] and [https://en.wikipedia.org/wiki/Homo_sapiens Homo sapiens]. The June 2004 RCSB PDB [https://pdb.rcsb.org/pdb/static.do?p=education_discussion/molecule_of_the_month/index.html Molecule of the Month] feature on ''Acetylcholinesterase'' by David S. Goodsell is [https://dx.doi.org/10.2210/rcsb_pdb/mom_2004_6 10.2210/rcsb_pdb/mom_2004_6]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1B41 OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=1B41 FirstGlance]. <br> | |||
</td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 2.76Å</td></tr> | |||
- | <tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=FUC:ALPHA-L-FUCOSE'>FUC</scene>, <scene name='pdbligand=NAG:N-ACETYL-D-GLUCOSAMINE'>NAG</scene></td></tr> | ||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1b41 FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1b41 OCA], [https://pdbe.org/1b41 PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1b41 RCSB], [https://www.ebi.ac.uk/pdbsum/1b41 PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1b41 ProSAT]</span></td></tr> | |||
</table> | |||
== Function == | |||
[https://www.uniprot.org/uniprot/ACES_HUMAN ACES_HUMAN] Terminates signal transduction at the neuromuscular junction by rapid hydrolysis of the acetylcholine released into the synaptic cleft. Role in neuronal apoptosis.<ref>PMID:2714437</ref> <ref>PMID:1748670</ref> <ref>PMID:1517212</ref> <ref>PMID:11985878</ref> | |||
== Evolutionary Conservation == | |||
[[Image:Consurf_key_small.gif|200px|right]] | |||
Check<jmol> | |||
<jmolCheckbox> | |||
<scriptWhenChecked>; select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/b4/1b41_consurf.spt"</scriptWhenChecked> | |||
<scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> | |||
<text>to colour the structure by Evolutionary Conservation</text> | |||
</jmolCheckbox> | |||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1b41 ConSurf]. | |||
<div style="clear:both"></div> | |||
<div style="background-color:#fffaf0;"> | |||
== Publication Abstract from PubMed == | |||
Structures of recombinant wild-type human acetylcholinesterase and of its E202Q mutant as complexes with fasciculin-II, a 'three-finger' polypeptide toxin purified from the venom of the eastern green mamba (Dendroaspis angusticeps), are reported. The structure of the complex of the wild-type enzyme was solved to 2.8 A resolution by molecular replacement starting from the structure of the complex of Torpedo californica acetylcholinesterase with fasciculin-II and verified by starting from a similar complex with mouse acetylcholinesterase. The overall structure is surprisingly similar to that of the T. californica enzyme with fasciculin-II and, as expected, to that of the mouse acetylcholinesterase complex. The structure of the E202Q mutant complex was refined starting from the corresponding wild-type human acetylcholinesterase structure, using the 2.7 A resolution data set collected. Comparison of the two structures shows that removal of the charged group from the protein core and its substitution by a neutral isosteric moiety does not disrupt the functional architecture of the active centre. One of the elements of this architecture is thought to be a hydrogen-bond network including residues Glu202, Glu450, Tyr133 and two bridging molecules of water, which is conserved in other vertebrate acetylcholinesterases as well as in the human enzyme. The present findings are consistent with the notion that the main role of this network is the proper positioning of the Glu202 carboxylate relative to the catalytic triad, thus defining its functional role in the interaction of acetylcholinesterase with substrates and inhibitors. | |||
Structures of recombinant native and E202Q mutant human acetylcholinesterase complexed with the snake-venom toxin fasciculin-II.,Kryger G, Harel M, Giles K, Toker L, Velan B, Lazar A, Kronman C, Barak D, Ariel N, Shafferman A, Silman I, Sussman JL Acta Crystallogr D Biol Crystallogr. 2000 Nov;56(Pt 11):1385-94. PMID:11053835<ref>PMID:11053835</ref> | |||
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | |||
</div> | |||
<div class="pdbe-citations 1b41" style="background-color:#fffaf0;"></div> | |||
= | |||
==See Also== | ==See Also== | ||
*[[Acetylcholinesterase|Acetylcholinesterase | *[[Acetylcholinesterase 3D structures|Acetylcholinesterase 3D structures]] | ||
*[[Fasciculin|Fasciculin]] | *[[Fasciculin|Fasciculin]] | ||
== References == | |||
== | <references/> | ||
< | __TOC__ | ||
</StructureSection> | |||
[[Category: Acetylcholinesterase]] | [[Category: Acetylcholinesterase]] | ||
[[Category: Dendroaspis angusticeps]] | [[Category: Dendroaspis angusticeps]] | ||
[[Category: Homo sapiens]] | [[Category: Homo sapiens]] | ||
[[Category: Large Structures]] | |||
[[Category: RCSB PDB Molecule of the Month]] | [[Category: RCSB PDB Molecule of the Month]] | ||
[[Category: Harel | [[Category: Harel M]] | ||
[[Category: Kryger | [[Category: Kryger G]] | ||
[[Category: Shafferman | [[Category: Shafferman A]] | ||
[[Category: Silman | [[Category: Silman I]] | ||
[[Category: Sussman | [[Category: Sussman JL]] | ||
Latest revision as of 08:36, 9 August 2023
HUMAN ACETYLCHOLINESTERASE COMPLEXED WITH FASCICULIN-II, GLYCOSYLATED PROTEINHUMAN ACETYLCHOLINESTERASE COMPLEXED WITH FASCICULIN-II, GLYCOSYLATED PROTEIN
Structural highlights
FunctionACES_HUMAN Terminates signal transduction at the neuromuscular junction by rapid hydrolysis of the acetylcholine released into the synaptic cleft. Role in neuronal apoptosis.[1] [2] [3] [4] Evolutionary Conservation Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. Publication Abstract from PubMedStructures of recombinant wild-type human acetylcholinesterase and of its E202Q mutant as complexes with fasciculin-II, a 'three-finger' polypeptide toxin purified from the venom of the eastern green mamba (Dendroaspis angusticeps), are reported. The structure of the complex of the wild-type enzyme was solved to 2.8 A resolution by molecular replacement starting from the structure of the complex of Torpedo californica acetylcholinesterase with fasciculin-II and verified by starting from a similar complex with mouse acetylcholinesterase. The overall structure is surprisingly similar to that of the T. californica enzyme with fasciculin-II and, as expected, to that of the mouse acetylcholinesterase complex. The structure of the E202Q mutant complex was refined starting from the corresponding wild-type human acetylcholinesterase structure, using the 2.7 A resolution data set collected. Comparison of the two structures shows that removal of the charged group from the protein core and its substitution by a neutral isosteric moiety does not disrupt the functional architecture of the active centre. One of the elements of this architecture is thought to be a hydrogen-bond network including residues Glu202, Glu450, Tyr133 and two bridging molecules of water, which is conserved in other vertebrate acetylcholinesterases as well as in the human enzyme. The present findings are consistent with the notion that the main role of this network is the proper positioning of the Glu202 carboxylate relative to the catalytic triad, thus defining its functional role in the interaction of acetylcholinesterase with substrates and inhibitors. Structures of recombinant native and E202Q mutant human acetylcholinesterase complexed with the snake-venom toxin fasciculin-II.,Kryger G, Harel M, Giles K, Toker L, Velan B, Lazar A, Kronman C, Barak D, Ariel N, Shafferman A, Silman I, Sussman JL Acta Crystallogr D Biol Crystallogr. 2000 Nov;56(Pt 11):1385-94. PMID:11053835[5] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences
|
| ||||||||||||||||||