Phospholipase A2: Difference between revisions
No edit summary |
No edit summary |
||
| (34 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
<StructureSection load='' size=' | <StructureSection load='' size='350' side='right' scene=Phospholipase_A2/Cv/1 caption='Phospholipase A2 complex with ethanol, phosphate and Ca+2 ion (green) (PDB code [[1yxh]])'> | ||
__TOC__ | |||
[[Phospholipase A2]] (PLA2) is an enzyme which releases fatty acids from glycerol. It is found in mammals and in snake venoms. PLA2 releases arachidonic acid from membranes causing inflammation and pain. PLA2 | == Function == | ||
[[Phospholipase A2]] (PLA2) is an enzyme which releases fatty acids from glycerol. It is found in mammals and in snake venoms<ref>PMID:8175726</ref>. PLA2 releases arachidonic acid from membranes causing inflammation and pain. The PLA2 contains many isozymes which are ordered by groups and named accordingly, ie., group I is PLA2G1.<br /> | |||
*'''PLA2 group II''' is induced in inflammation and present in atherosclerotic lesions<ref>PMID:10323782</ref>. <br /> | |||
*'''PLA2 group V''' is active in leukocytes recruitment<ref>PMID:20232296</ref>. <br /> | |||
*'''PLA2 group X''' regulates cysteinyl leukotriene synthesis<ref>PMID:26139511</ref>. <br /> | |||
*'''PLA2 group XV''' is located in the endocytic system and may be involved in CD1d function<ref>PMID:23943550</ref>. <br /> | |||
*'''PLA2 group XVI''' is adipose-specific<ref>PMID:18614531</ref>. <br /> | |||
*'''Lipoprotein-associated phospholipase A2''' or '''platelet-activating factor acetylhydrolase''' (Lp-PLA2) degrades platelet-activating factor and oxidized phospholipids into inactive metabolites. Platelet-activating factor is a potent phospholipid activator and mediator of inflammation, platelet aggregation and other leukocyte functions<ref>PMID:7700381</ref>. <br />. | |||
*'''Bothropstoxin (BTX)''' is PLA2 from the snake ''Bothrops jararacussu''.<br /> | |||
*'''Piratoxin (PTX)''' is PLA2 from the snake ''Bothrops piraña''.<br /> | |||
*'''Viperotoxin (VTX)''' is a heterodimer of a very homologous PLA2 called '''RV-4/RV-7'''.<br /> | |||
*'''Cystolic PLA2 (cPLA2)''' is intracellular. It is larger than the secreted PLA2 and contains the targeting C2 domain. <br /> | |||
*'''Pro-phosphlipase (PPLA2)''' is a pancreatic PLA2 whose 7-mer N-terminal peptide is cleaved off to produce the active PLA2.<br /> | |||
*For details on Lys49 snake-venom PLA2 see [[Anum-II]].<br /> | |||
*See also [[Phospholipase (Hebrew)]]. | |||
== Relevance == | |||
PLA2 serve as pharmacological targets for therapeutical treatment of diseases like atherosclerosis, immune disorders, cardiovascular diseases and cancer<ref>PMID:24907600</ref>. Reduced Lp-PLA2 activity is observed in patients with severe sepsis. There is association between the level of Lp-PLA2 in plasma and the risk of future cardiovascular events<ref>PMID:19272461</ref>. | |||
< | ==Diclofenac binding to Phospholipase A2<ref>PMID:16552142</ref>== | ||
'''Abstract from Pubmed''' | |||
Type IIA secretory phospholipase A2 (PLA2) enzymes catalyze the hydrolysis of the sn-2 ester bond of glycerophospholipids to release fatty acids and lysophospholipids. In order to elucidate the role of PLA2 in inflammatory disorders and to determine the mode of binding of non-steroidal anti-inflammatory drugs (NSAIDs) to PLA2, the detailed three-dimensional structure of a complex formed between a group IIA PLA2 from Daboia russelli pulchella and 2-[(2,6-dichlorophenyl)amino]benzeneacetic acid (diclofenac) has been determined. The preformed complex was crystallized by equilibrating the protein solution against a mixture of 0.20 M ammonium sulfate and 30% PEG 4000. The crystals belong to space group P4(3), with unit-cell parameters a = b = 53.0, c = 48.4 A. The structure was solved by the molecular-replacement method and refined to R(cryst) and R(free) factors of 0.192 and 0.211, respectively, using reflections to 2.7 A resolution. The structure showed that diclofenac occupies a very favourable position in the centre of the substrate-binding hydrophobic channel that allows a number of intermolecular interactions. The binding mode of diclofenac involved crucial interactions with important residues for substrate recognition such as Asp49, His48 and Gly30. In addition, it included three new interactions involving its Cl atoms with Phe5, Ala18 and Tyr22. It also showed an extensive network of hydrophobic interactions involving almost all of the residues of the substrate-binding hydrophobic channel. The binding affinity of diclofenac was determined using surface plasmon resonance, which gave an equilibrium constant of 4.8 +/- 0.2 x 10(-8) M. | |||
The <scene name='Diclofenac_binding_to_Phospholipase_A2/Pla2-diclofenac/1'>overall structure of the complex</scene> | |||
shows three mail helices in phospholipase A2. | |||
The <scene name='Diclofenac_binding_to_Phospholipase_A2/Active_site/2'>active site residues</scene> are His 48, Asp 49, Tyr 52 and Glu 99 in the structure. | |||
Diclofenac makes several <scene name='Diclofenac_binding_to_Phospholipase_A2/Hydro/1'>Hydrophobic interactions</scene> with the substrate binding site of enzyme. ligand binding is shown in <scene name='Diclofenac_binding_to_Phospholipase_A2/Space_filling/1'>space filling</scene> model of the complex. See also [[Diclofenac]]. | |||
== | == Crystal structure of porcine pancreatic phospholipase A<sub>2</sub> in complex with 2-methoxycyclohexa-2-5-diene-1,4-dione == | ||
<ref >DOI 10.1080/21553769.2012.689262</ref> | |||
<scene name='Journal:FLS:1/Cv/1'>Crystal structure of porcine pancreatic phospholipase A2</scene> | |||
=== | <scene name='Journal:FLS:1/Cv/4'>Curcumin</scene> possesses anti-inflammatory activity. The binding of curcumin with PLA<sub>2</sub> was studied using X-ray crystallography. Since the electron density found in the active site did not match with curcumin, <scene name='Journal:FLS:1/Cv/5'>2-methoxycyclohexa-2-5-diene-1,4-dione (MCW)</scene> (the photo-degraded product of curcumin) <scene name='Journal:FLS:1/Cv/6'>was fitted</scene> in the unexplained electron density. To understand the <scene name='Journal:FLS:1/Cv/9'>binding mode of actual curcumin</scene>, molecular docking studies was carried out. <scene name='Journal:FLS:1/Cv/10'>Both crystallographic and docked structures were superimposed</scene> with respect to the ligand position and identified that <scene name='Journal:FLS:1/Cv/13'>curcumin is binding in the hydrophobic cavity</scene> of PLA<sub>2</sub> with a binding energy -16.81 Kcal/mol. The binding mode is in such a manner that it can prevent the entry of substrate to the hydrophobic active site. These studies indicate that curcumin can be act as an inhibitor to PLA<sub>2</sub>. | ||
== '''Interaction of Atropine with Phospholipase A2''' == | |||
= | <scene name='42/420811/Cv/1'>Atropine in complex with phospholipase A2</scene> ([[1th6]]). | ||
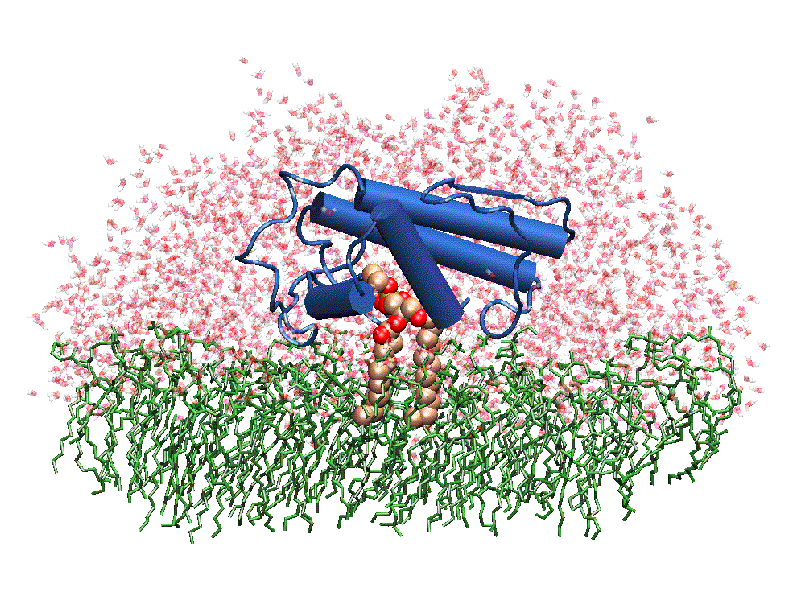
[[ | [[Image:Phospholipase A2.gif|thumb|left|350px|Phospholipase 2A in complex with cell membrane]] | ||
{{Clear}} | |||
In addition to its ability to form complexes with acetylcholine receptors, atropine can also complex with phospholipase A2. Phospholipase A2 is a category of heat-stable enzymes which are involved in cell signaling processes, such as the inflammatory response. <ref>Kumar, Jainendra; Bala, Priti; Vihwal, Preeti. ''Analysis of Interaction of atropine with phospholipase A2 (1th6.pdb)''. Department of Botany and Biotechnlogy, College of Commerce, Patna, India.</ref>. Phospholipase 2A is an upstream regulator of inflammatory processes, and more specifically, it recognizes the sn-2 acyl bond of phospholipids and catalytically hydrolyzes the bond, releasing lysophospholipids <ref> Phospholipase A2. http://www.worldlingo.com/ma/enwiki/en/Phospholipase_A2 </ref>. | |||
This protein is found in mammals, reptile venom, and bacteria. In humans, the overproduction of phospholipase A2 leads to neurologic disorders such as schizophrenia and possibly autism <ref> Phospholipase A2. http://www.worldlingo.com/ma/enwiki/en/Phospholipase_A2 </ref>. An inhibitor of Phospholipase A2, such as Atropine, could be used to treat disorders associated with neural trauma, since Phospholipase A2 increases inflammation which could be potentially complicate neural trauma cases <ref> Phospholipase A2. http://www.worldlingo.com/ma/enwiki/en/Phospholipase_A2 </ref>. | |||
The image to the above shows the membrane-bound phospholipase A2 in blue <ref> pla2. http://www.ks.uiuc.edu/Research/smd_imd/pla2/pla2.gif </ref>. | |||
=== | === '''Atropine in the Active Site of Phospholipase A2''' === | ||
Atropine is an inhibitor of phospholipase 2A, and can be seen in complex with this enzyme on the left. The <scene name='Sandbox_53/Atropine_structure/1'>structure of atropine</scene> can be seen more clearly in gray using the ball-and stick representation of the drug and protein. It can also be seen in green in this <scene name='Sandbox_53/Phospholipase2a_composition/1'>space-filling model</scene>, where protein appears in brown, ligands appear in green, and solvents appear in blue. Finally, the | |||
<scene name='Sandbox_53/Phospholipase2a_rainbow/1'>N to C terminal</scene> portions of the protein can be highlighted from blue to red in a rainbow, and the active site with atropine can be seen in the middle of the protein. | |||
== | Atropine interacts with phospholipase 2A at residues asp29 and tyr49 on the protein. The | ||
<scene name='Sandbox_53/Phospholipase2a_residues/1'>residues</scene> of atropine interacting with phospholipase 2A can be seen on the right. The amino acid residues in the active site are labeled. As seen in the acetylcholine receptor, the <scene name='Sandbox_53/Phospholipase_hyrophobic/1'>hydrophobic</scene> regions of the phospholipase 2A enzyme are found in the active site, which is where the atropine binds and inhibits the enzyme. The hydrophobic regions, represented in gray, can be seen surrounding atropine, which is positioned in the active site and capped by red oxygen atoms. | |||
Removing the labels, atropine can be seen making contact with the atoms emphasized by the space filling model, interacting with the <scene name='Sandbox_53/Phospholipase2a_interactions/1'>active site</scene> of phospholipase 2A through white as-tricks. | |||
== | == 3D Structures of Phospholipase A2 == | ||
[[Phospholipase A2 3D structures]] | |||
</StructureSection> | |||
== | == References == | ||
<references/> | <references/> | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Latest revision as of 13:53, 4 July 2023
FunctionPhospholipase A2 (PLA2) is an enzyme which releases fatty acids from glycerol. It is found in mammals and in snake venoms[1]. PLA2 releases arachidonic acid from membranes causing inflammation and pain. The PLA2 contains many isozymes which are ordered by groups and named accordingly, ie., group I is PLA2G1.
RelevancePLA2 serve as pharmacological targets for therapeutical treatment of diseases like atherosclerosis, immune disorders, cardiovascular diseases and cancer[8]. Reduced Lp-PLA2 activity is observed in patients with severe sepsis. There is association between the level of Lp-PLA2 in plasma and the risk of future cardiovascular events[9]. Diclofenac binding to Phospholipase A2[10]Abstract from Pubmed Type IIA secretory phospholipase A2 (PLA2) enzymes catalyze the hydrolysis of the sn-2 ester bond of glycerophospholipids to release fatty acids and lysophospholipids. In order to elucidate the role of PLA2 in inflammatory disorders and to determine the mode of binding of non-steroidal anti-inflammatory drugs (NSAIDs) to PLA2, the detailed three-dimensional structure of a complex formed between a group IIA PLA2 from Daboia russelli pulchella and 2-[(2,6-dichlorophenyl)amino]benzeneacetic acid (diclofenac) has been determined. The preformed complex was crystallized by equilibrating the protein solution against a mixture of 0.20 M ammonium sulfate and 30% PEG 4000. The crystals belong to space group P4(3), with unit-cell parameters a = b = 53.0, c = 48.4 A. The structure was solved by the molecular-replacement method and refined to R(cryst) and R(free) factors of 0.192 and 0.211, respectively, using reflections to 2.7 A resolution. The structure showed that diclofenac occupies a very favourable position in the centre of the substrate-binding hydrophobic channel that allows a number of intermolecular interactions. The binding mode of diclofenac involved crucial interactions with important residues for substrate recognition such as Asp49, His48 and Gly30. In addition, it included three new interactions involving its Cl atoms with Phe5, Ala18 and Tyr22. It also showed an extensive network of hydrophobic interactions involving almost all of the residues of the substrate-binding hydrophobic channel. The binding affinity of diclofenac was determined using surface plasmon resonance, which gave an equilibrium constant of 4.8 +/- 0.2 x 10(-8) M. The shows three mail helices in phospholipase A2. The are His 48, Asp 49, Tyr 52 and Glu 99 in the structure. Diclofenac makes several with the substrate binding site of enzyme. ligand binding is shown in model of the complex. See also Diclofenac. Crystal structure of porcine pancreatic phospholipase A2 in complex with 2-methoxycyclohexa-2-5-diene-1,4-dione
possesses anti-inflammatory activity. The binding of curcumin with PLA2 was studied using X-ray crystallography. Since the electron density found in the active site did not match with curcumin, (the photo-degraded product of curcumin) in the unexplained electron density. To understand the , molecular docking studies was carried out. with respect to the ligand position and identified that of PLA2 with a binding energy -16.81 Kcal/mol. The binding mode is in such a manner that it can prevent the entry of substrate to the hydrophobic active site. These studies indicate that curcumin can be act as an inhibitor to PLA2. Interaction of Atropine with Phospholipase A2(1th6).  In addition to its ability to form complexes with acetylcholine receptors, atropine can also complex with phospholipase A2. Phospholipase A2 is a category of heat-stable enzymes which are involved in cell signaling processes, such as the inflammatory response. [12]. Phospholipase 2A is an upstream regulator of inflammatory processes, and more specifically, it recognizes the sn-2 acyl bond of phospholipids and catalytically hydrolyzes the bond, releasing lysophospholipids [13]. This protein is found in mammals, reptile venom, and bacteria. In humans, the overproduction of phospholipase A2 leads to neurologic disorders such as schizophrenia and possibly autism [14]. An inhibitor of Phospholipase A2, such as Atropine, could be used to treat disorders associated with neural trauma, since Phospholipase A2 increases inflammation which could be potentially complicate neural trauma cases [15]. The image to the above shows the membrane-bound phospholipase A2 in blue [16]. Atropine in the Active Site of Phospholipase A2Atropine is an inhibitor of phospholipase 2A, and can be seen in complex with this enzyme on the left. The can be seen more clearly in gray using the ball-and stick representation of the drug and protein. It can also be seen in green in this , where protein appears in brown, ligands appear in green, and solvents appear in blue. Finally, the portions of the protein can be highlighted from blue to red in a rainbow, and the active site with atropine can be seen in the middle of the protein. Atropine interacts with phospholipase 2A at residues asp29 and tyr49 on the protein. The of atropine interacting with phospholipase 2A can be seen on the right. The amino acid residues in the active site are labeled. As seen in the acetylcholine receptor, the regions of the phospholipase 2A enzyme are found in the active site, which is where the atropine binds and inhibits the enzyme. The hydrophobic regions, represented in gray, can be seen surrounding atropine, which is positioned in the active site and capped by red oxygen atoms. Removing the labels, atropine can be seen making contact with the atoms emphasized by the space filling model, interacting with the of phospholipase 2A through white as-tricks. 3D Structures of Phospholipase A2Phospholipase A2 3D structures
|
| ||||||||||
ReferencesReferences
- ↑ Dennis EA. Diversity of group types, regulation, and function of phospholipase A2. J Biol Chem. 1994 May 6;269(18):13057-60. PMID:8175726
- ↑ Leitinger N, Watson AD, Hama SY, Ivandic B, Qiao JH, Huber J, Faull KF, Grass DS, Navab M, Fogelman AM, de Beer FC, Lusis AJ, Berliner JA. Role of group II secretory phospholipase A2 in atherosclerosis: 2. Potential involvement of biologically active oxidized phospholipids. Arterioscler Thromb Vasc Biol. 1999 May;19(5):1291-8. PMID:10323782
- ↑ Lapointe S, Brkovic A, Cloutier I, Tanguay JF, Arm JP, Sirois MG. Group V secreted phospholipase A2 contributes to LPS-induced leukocyte recruitment. J Cell Physiol. 2010 Jul;224(1):127-34. doi: 10.1002/jcp.22106. PMID:20232296 doi:http://dx.doi.org/10.1002/jcp.22106
- ↑ Hallstrand TS, Lai Y, Hooper KA, Oslund RC, Altemeier WA, Matute-Bello G, Gelb MH. Endogenous secreted phospholipase A2 group X regulates cysteinyl leukotrienes synthesis by human eosinophils. J Allergy Clin Immunol. 2016 Jan;137(1):268-77.e8. doi:, 10.1016/j.jaci.2015.05.026. Epub 2015 Jun 30. PMID:26139511 doi:http://dx.doi.org/10.1016/j.jaci.2015.05.026
- ↑ Platt RW, Brookhart MA, Cole SR, Westreich D, Schisterman EF. Reply to taguri and matsuyama. Stat Med. 2013 Sep 10;32(20):3592-3. doi: 10.1002/sim.5805. PMID:23943550 doi:http://dx.doi.org/10.1002/sim.5805
- ↑ Duncan RE, Sarkadi-Nagy E, Jaworski K, Ahmadian M, Sul HS. Identification and functional characterization of adipose-specific phospholipase A2 (AdPLA). J Biol Chem. 2008 Sep 12;283(37):25428-36. doi: 10.1074/jbc.M804146200. Epub 2008, Jul 9. PMID:18614531 doi:http://dx.doi.org/10.1074/jbc.M804146200
- ↑ Tjoelker LW, Wilder C, Eberhardt C, Stafforini DM, Dietsch G, Schimpf B, Hooper S, Le Trong H, Cousens LS, Zimmerman GA, Yamada Y, McIntyre TM, Prescott SM, Gray PW. Anti-inflammatory properties of a platelet-activating factor acetylhydrolase. Nature. 1995 Apr 6;374(6522):549-53. PMID:7700381 doi:http://dx.doi.org/10.1038/374549a0
- ↑ Quach ND, Arnold RD, Cummings BS. Secretory phospholipase A2 enzymes as pharmacological targets for treatment of disease. Biochem Pharmacol. 2014 Aug 15;90(4):338-48. doi: 10.1016/j.bcp.2014.05.022. Epub, 2014 Jun 4. PMID:24907600 doi:http://dx.doi.org/10.1016/j.bcp.2014.05.022
- ↑ Tellis CC, Tselepis AD. The role of lipoprotein-associated phospholipase A2 in atherosclerosis may depend on its lipoprotein carrier in plasma. Biochim Biophys Acta. 2009 May;1791(5):327-38. doi: 10.1016/j.bbalip.2009.02.015. PMID:19272461 doi:http://dx.doi.org/10.1016/j.bbalip.2009.02.015
- ↑ Singh N, Jabeen T, Sharma S, Somvanshi RK, Dey S, Srinivasan A, Singh TP. Specific binding of non-steroidal anti-inflammatory drugs (NSAIDs) to phospholipase A2: structure of the complex formed between phospholipase A2 and diclofenac at 2.7 A resolution. Acta Crystallogr D Biol Crystallogr. 2006 Apr;62(Pt 4):410-6. Epub 2006, Mar 18. PMID:16552142 doi:10.1107/S0907444906003660
- ↑ Crystal structure of porcine pancreatic phospholipase a2 in complex with 2-methoxycyclohexa-2-5-diene-1,4-dione. Dileep KV, Tintu I, Mandal PK, Karthe P, Haridas M, Sadasivan C. Frontiers In Life Sci. (2012) doi:http://dx.doi.org/10.1080/21553769.2012.689262
- ↑ Kumar, Jainendra; Bala, Priti; Vihwal, Preeti. Analysis of Interaction of atropine with phospholipase A2 (1th6.pdb). Department of Botany and Biotechnlogy, College of Commerce, Patna, India.
- ↑ Phospholipase A2. http://www.worldlingo.com/ma/enwiki/en/Phospholipase_A2
- ↑ Phospholipase A2. http://www.worldlingo.com/ma/enwiki/en/Phospholipase_A2
- ↑ Phospholipase A2. http://www.worldlingo.com/ma/enwiki/en/Phospholipase_A2
- ↑ pla2.