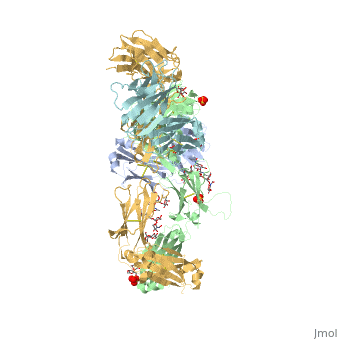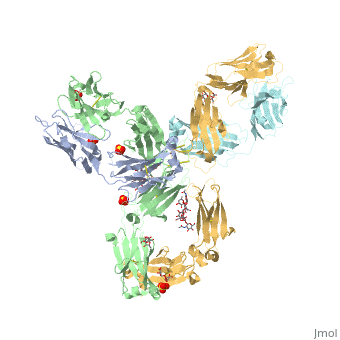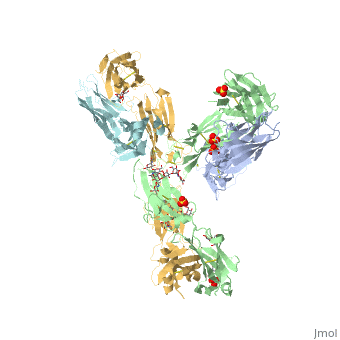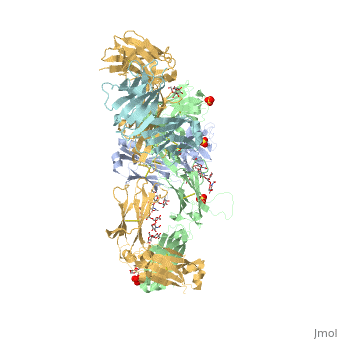5dk3: Difference between revisions
No edit summary |
No edit summary |
||
| (3 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
==Crystal Structure of Pembrolizumab, a full length IgG4 antibody== | ==Crystal Structure of Pembrolizumab, a full length IgG4 antibody== | ||
<StructureSection load='5dk3' size='340' side='right' caption='[[5dk3]], [[Resolution|resolution]] 2.28Å' scene=''> | <StructureSection load='5dk3' size='340' side='right'caption='[[5dk3]], [[Resolution|resolution]] 2.28Å' scene=''> | ||
== Structural highlights == | == Structural highlights == | ||
<table><tr><td colspan='2'>[[5dk3]] is a 4 chain structure. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=5DK3 OCA]. For a <b>guided tour on the structure components</b> use [ | <table><tr><td colspan='2'>[[5dk3]] is a 4 chain structure with sequence from [https://en.wikipedia.org/wiki/Homo_sapiens Homo sapiens]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=5DK3 OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=5DK3 FirstGlance]. <br> | ||
</td></tr><tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat"><scene name='pdbligand=BMA:BETA-D-MANNOSE'>BMA</scene>, <scene name='pdbligand=FUC:ALPHA-L-FUCOSE'>FUC</scene>, <scene name='pdbligand=MAN:ALPHA-D-MANNOSE'>MAN</scene>, <scene name='pdbligand=NAG:N-ACETYL-D-GLUCOSAMINE'>NAG</scene>, <scene name='pdbligand= | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 2.28Å</td></tr> | ||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[ | <tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=BMA:BETA-D-MANNOSE'>BMA</scene>, <scene name='pdbligand=FRU:FRUCTOSE'>FRU</scene>, <scene name='pdbligand=FUC:ALPHA-L-FUCOSE'>FUC</scene>, <scene name='pdbligand=GLC:ALPHA-D-GLUCOSE'>GLC</scene>, <scene name='pdbligand=MAN:ALPHA-D-MANNOSE'>MAN</scene>, <scene name='pdbligand=NAG:N-ACETYL-D-GLUCOSAMINE'>NAG</scene>, <scene name='pdbligand=PRD_900003:sucrose'>PRD_900003</scene>, <scene name='pdbligand=SO4:SULFATE+ION'>SO4</scene></td></tr> | ||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=5dk3 FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=5dk3 OCA], [https://pdbe.org/5dk3 PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=5dk3 RCSB], [https://www.ebi.ac.uk/pdbsum/5dk3 PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=5dk3 ProSAT]</span></td></tr> | |||
</table> | </table> | ||
<div style="background-color:#fffaf0;"> | |||
== Publication Abstract from PubMed == | |||
Immunoglobulin G4 antibodies exhibit unusual properties with important biological consequences. We report the structure of the human full-length IgG4 S228P anti-PD1 antibody pembrolizumab, solved to 2.3-A resolution. Pembrolizumab is a compact molecule, consistent with the presence of a short hinge region. The Fc domain is glycosylated at the CH2 domain on both chains, but one CH2 domain is rotated 120 degrees with respect to the conformation observed in all reported structures to date, and its glycan chain faces the solvent. We speculate that this new conformation is driven by the shorter hinge. The structure suggests a role for the S228P mutation in preventing the IgG4 arm exchange. In addition, this unusual Fc conformation suggests possible structural diversity between IgG subclasses and shows that use of isolated antibody fragments could mask potentially important interactions, owing to molecular flexibility. | |||
Structure of full-length human anti-PD1 therapeutic IgG4 antibody pembrolizumab.,Scapin G, Yang X, Prosise WW, McCoy M, Reichert P, Johnston JM, Kashi RS, Strickland C Nat Struct Mol Biol. 2015 Dec;22(12):953-8. doi: 10.1038/nsmb.3129. Epub 2015 Nov, 23. PMID:26595420<ref>PMID:26595420</ref> | |||
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | |||
</div> | |||
<div class="pdbe-citations 5dk3" style="background-color:#fffaf0;"></div> | |||
==See Also== | |||
*[[Monoclonal Antibodies 3D structures|Monoclonal Antibodies 3D structures]] | |||
== References == | |||
<references/> | |||
__TOC__ | __TOC__ | ||
</StructureSection> | </StructureSection> | ||
[[Category: | [[Category: Homo sapiens]] | ||
[[Category: | [[Category: Large Structures]] | ||
[[Category: | [[Category: Prosise W]] | ||
[[Category: | [[Category: Reichert P]] | ||
[[Category: | [[Category: Scapin G]] | ||
Latest revision as of 00:45, 29 June 2023
Crystal Structure of Pembrolizumab, a full length IgG4 antibodyCrystal Structure of Pembrolizumab, a full length IgG4 antibody
Structural highlights
Publication Abstract from PubMedImmunoglobulin G4 antibodies exhibit unusual properties with important biological consequences. We report the structure of the human full-length IgG4 S228P anti-PD1 antibody pembrolizumab, solved to 2.3-A resolution. Pembrolizumab is a compact molecule, consistent with the presence of a short hinge region. The Fc domain is glycosylated at the CH2 domain on both chains, but one CH2 domain is rotated 120 degrees with respect to the conformation observed in all reported structures to date, and its glycan chain faces the solvent. We speculate that this new conformation is driven by the shorter hinge. The structure suggests a role for the S228P mutation in preventing the IgG4 arm exchange. In addition, this unusual Fc conformation suggests possible structural diversity between IgG subclasses and shows that use of isolated antibody fragments could mask potentially important interactions, owing to molecular flexibility. Structure of full-length human anti-PD1 therapeutic IgG4 antibody pembrolizumab.,Scapin G, Yang X, Prosise WW, McCoy M, Reichert P, Johnston JM, Kashi RS, Strickland C Nat Struct Mol Biol. 2015 Dec;22(12):953-8. doi: 10.1038/nsmb.3129. Epub 2015 Nov, 23. PMID:26595420[1] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences
|
| ||||||||||||||||||