Dihydrodipicolinate synthase: Difference between revisions
Michal Harel (talk | contribs) No edit summary |
Michal Harel (talk | contribs) No edit summary |
||
| (18 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
<StructureSection load=' | <StructureSection load='2v9d' size='350' side='right' caption='Structure of Se-Met dihydrodipicolinate synthase (PDB entry [[2v9d]])' scene=''> | ||
[[ | |||
'''CRYSTAL STRUCTURE OF YAGE, A PROPHAGE PROTEIN BELONGING TO THE DIHYDRODIPICOLINIC ACID SYNTHASE FAMILY FROM ''E. COLI K12'''''<br /> | '''CRYSTAL STRUCTURE OF YAGE, A PROPHAGE PROTEIN BELONGING TO THE DIHYDRODIPICOLINIC ACID SYNTHASE FAMILY FROM ''E. COLI K12'''''<br /> | ||
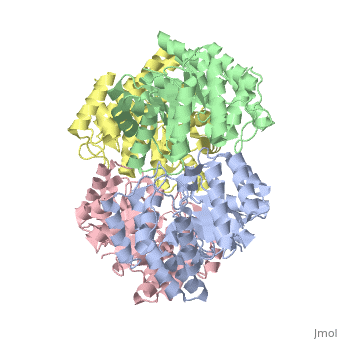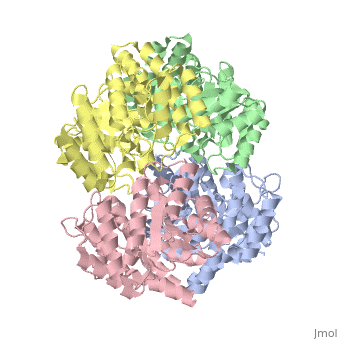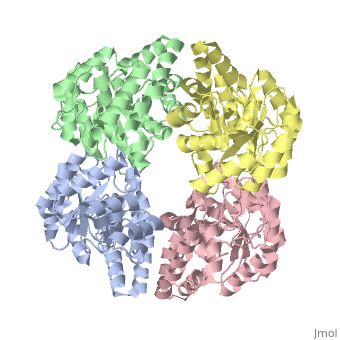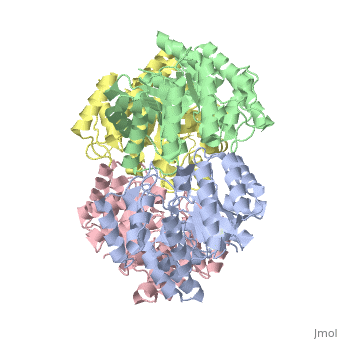
One approach of the incorporation of genetic material from different species into bacterial genomes is infection by a bacteriophage. The ''yagE'' gene is located in the ''E. coli K12'' genome and it is part of prophage ''CP4-6'' encoding a 33-kDa putative dihydrodipicolinate synthase (DHDPS)-like protein (UniProtKB/Swiss-prot: [http://www.expasy.org/uniprot/P75682 P75682]). This DHDPS-like domain is a member of the N-acetyl neuraminate lyase (NAL) subfamily comprises an 8-fold <scene name='2v9d/Alpha_beta/2'>α/β-barrel</scene> ([http://en.wikipedia.org/wiki/TIM_barrel TIM barrel]) with a small C-terminal α-helical domain (Inter pro: [http://www.ebi.ac.uk/interpro/ISearch?query=IPR005263 IPR005263]). Many enzymes possess this DHDPS domain (''e.g.'' HBPHA, KDGA, NAL, DOGDH, and DHDPS). All these enzymes contain an 8-fold α/β barrel and a small C-terminal α-helical region. <scene name='2v8z/Comparison/2'>Comparison</scene> between <font color='red'><b>orthorhombic</b></font> ([[2v8z]]) and <font color='cyan'><b>monoclinic</b></font> ([[2v9d]]) crystal forms of YagE revealed striking similarity. | One approach of the incorporation of genetic material from different species into bacterial genomes is infection by a bacteriophage. The ''yagE'' gene is located in the ''E. coli K12'' genome and it is part of prophage ''CP4-6'' encoding a 33-kDa putative '''dihydrodipicolinate synthase''' or '''4-hydroxy-tetrahydrodihydrodipicolinate synthase''' (DHDPS)-like protein (UniProtKB/Swiss-prot: [http://www.expasy.org/uniprot/P75682 P75682]). This DHDPS-like domain is a member of the N-acetyl neuraminate lyase (NAL) subfamily comprises an 8-fold <scene name='2v9d/Alpha_beta/2'>α/β-barrel</scene> ([http://en.wikipedia.org/wiki/TIM_barrel TIM barrel]) with a small C-terminal α-helical domain (Inter pro: [http://www.ebi.ac.uk/interpro/ISearch?query=IPR005263 IPR005263]). Many enzymes possess this DHDPS domain (''e.g.'' HBPHA, KDGA, NAL, DOGDH, and DHDPS). All these enzymes contain an 8-fold α/β barrel and a small C-terminal α-helical region. <scene name='2v8z/Comparison/2'>Comparison</scene> between <font color='red'><b>orthorhombic</b></font> ([[2v8z]]) and <font color='cyan'><b>monoclinic</b></font> ([[2v9d]]) crystal forms of YagE revealed striking similarity. | ||
{{Clear}} | {{Clear}} | ||
| Line 30: | Line 12: | ||
Members of the NAL protein subfamily have very similar active sites and a single amino acid substitution can significantly change their function. For example, NAL ([[1nal]]) gets DHDPS activity by substitution of a <scene name='2v9d/Leucine142/2'>leucine</scene> <font color='orange'><b>(orange)</b></font> to <scene name='2v9d/Arginine142/2'>arginine</scene> <font color='blue'><b>(blue)</b></font> at position 142. The possible active site region of <font color='cyan'><b>YagE</b></font> demonstrates closest sequence similarity to the active site of <font color='magenta'><b>KDG aldolase of SsKDGA</b></font> ([[1w3i]]) and <font color='black'><b>NAL of EcNAL</b></font> ([[1nal]]) (yellow), suggesting that this protein can perform either of these functions. Although the active site of EcDHDPS ([[1xky]]) and BaDHDPS ([[2ats]]) shows similarities, the important residue that differentiates between NAL and DHDPS, namely Leu142 (in [[1nal]]), is also present in YagE <scene name='2v9d/Leucine150/2'>(Leu150)</scene> <font color='cyan'><b>(labeled cyan)</b></font> at that particular position suggesting that YagE performs a NAL-related function rather than DHDPS-related one. In conclusion, the high-resolution X-ray structure of YagE provides a clue that it probably belongs to the NAL subfamily of proteins. Although the exact molecular function of YagE is still unknown, its structure provides a handle for understanding its molecular function based on knowledge about conserved residues of the putative active site. | Members of the NAL protein subfamily have very similar active sites and a single amino acid substitution can significantly change their function. For example, NAL ([[1nal]]) gets DHDPS activity by substitution of a <scene name='2v9d/Leucine142/2'>leucine</scene> <font color='orange'><b>(orange)</b></font> to <scene name='2v9d/Arginine142/2'>arginine</scene> <font color='blue'><b>(blue)</b></font> at position 142. The possible active site region of <font color='cyan'><b>YagE</b></font> demonstrates closest sequence similarity to the active site of <font color='magenta'><b>KDG aldolase of SsKDGA</b></font> ([[1w3i]]) and <font color='black'><b>NAL of EcNAL</b></font> ([[1nal]]) (yellow), suggesting that this protein can perform either of these functions. Although the active site of EcDHDPS ([[1xky]]) and BaDHDPS ([[2ats]]) shows similarities, the important residue that differentiates between NAL and DHDPS, namely Leu142 (in [[1nal]]), is also present in YagE <scene name='2v9d/Leucine150/2'>(Leu150)</scene> <font color='cyan'><b>(labeled cyan)</b></font> at that particular position suggesting that YagE performs a NAL-related function rather than DHDPS-related one. In conclusion, the high-resolution X-ray structure of YagE provides a clue that it probably belongs to the NAL subfamily of proteins. Although the exact molecular function of YagE is still unknown, its structure provides a handle for understanding its molecular function based on knowledge about conserved residues of the putative active site. | ||
{{Clear}} | {{Clear}} | ||
</StructureSection> | |||
==3D structures of dihydrodipicolinate synthase== | |||
Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | |||
{{#tree:id=OrganizedByTopic|openlevels=0| | |||
*Dihydrodipicolinate synthase or 4-hydroxy-tetrahydrodipicolinate synthase | |||
**[[4dpp]], [[6vvi]] – AtDPS – ''Arabidopsis thaliana''<br /> | |||
**[[3tuu]] – grDPS – grapevine<br /> | |||
**[[1dhp]], [[1s5t]], [[1s5v]], [[1s5w]], [[2a6l]], [[2a6n]], [[2ojp]], [[2pur]], [[3den]], [[3i7q]], [[3i7r]], [[7jz7]] – EcDPS (mutant) – ''Escherichia coli''<br /> | |||
**[[1yxc]] - EcDPS<br /> | |||
**[[1o5k]] – TmDPS – ''Thermotoga maritima''<br /> | |||
**[[3pb0]], [[3pb2]] – TmDPS (mutant)<br /> | |||
**[[1xky]], [[1xl9]] – BaDPS – ''Bacillus anthracis''<br /> | |||
**[[1xxx]] – MtDPS – ''Mycobacterium tuberculosis''<br /> | |||
**[[3l21]] – MtDPS (mutant)<br /> | |||
**[[2hmc]], [[2r8w]], [[3b4u]], [[4i7u]], [[4i7v]] – DPS – ''Agrobacterium tumdefaciens''<br /> | |||
**[[2ehh]] – DPS – ''Aquifex aeolicus''<br /> | |||
**[[2rfg]] – DPS – ''Hahella chejuensis''<br /> | |||
**[[2yxg]] – DPS – ''Methanocaldococcus jannaschii''<br /> | |||
**[[3d0c]] – DPS – ''Oceanobacillus iheyensis''<br /> | |||
**[[3daq]], [[3di0]] – SaDPS – ''Staphylococcus aureus''<br /> | |||
**[[3dz1]] – DPS – ''Rhodopseudomonas palustris''<br /> | |||
**[[3e96]] – DPS – ''Bacillus clausii''<br /> | |||
**[[3cpr]] – DPS – ''Corynebacterium glutamicum''<br /> | |||
**[[3g0s]] – DPS – ''Salmonella typhimurium''<br /> | |||
**[[3flu]] – DPS – ''Neisseria meningitides''<br /> | |||
**[[3ird]], [[3a5f]] – DPS – ''Clostridium botulinum''<br /> | |||
**[[3vfl]] – SpDPS – ''Streptococcus pneumoniae''<br /> | |||
**[[3ler]], [[3m5v]] – DPS – ''Campylobacter jejuni''<br /> | |||
**[[3noe]], [[3ps7]], [[3qze]] – PaDPS – ''Psuedomonas aeruginosa''<br /> | |||
**[[6p90]] - PaDPS (mutant) <br /> | |||
**[[3pud]], [[3pul]], [[4dxv]] – AbDPS – ''Acinetobacter baumannii''<br /> | |||
**[[3si9]] – DSP – ''Bartonella henselae''<br /> | |||
**[[4icn]] - SbDPS – ''Shewanella benthica''<br /> | |||
**[[4xky]] - DPS – ''Bacterioides thetaiotaomicron''<br /> | |||
**[[5czj]] - SmDPS – ''Sinorhizobium meliloti''<br /> | |||
**[[5ui3]] - DPS – ''Chlamydomonas reinhardtii''<br /> | |||
**[[7loy]] - ClDPS – ''Candidatus liberibacter''<br /> | |||
**[[6ue0]] - DPS – ''Klebsiella pneumoniae''<br /> | |||
**[[5ktl]] - DPS – ''Trichormus variabilis''<br /> | |||
**[[4r53]] - CjDPS – ''Campylobacter jejuni''<br /> | |||
*DHDPS binary complexes | |||
**[[4dpq]] - AtDPS + S-Lys<br /> | |||
**[[4i7w]], [[6vvh]] - AtDPS + Lys<br /> | |||
**[[7mds]] - AtDPS + inhibitor<br /> | |||
**[[4hnn]] - grDPS + Lys<br /> | |||
**[[5ud6]] - DPS + Lys – red alga<br /> | |||
**[[3pue]] - AbDPS + Lys<br /> | |||
**[[3tce]] - AbDPS + hydroxylysine<br /> | |||
**[[3tdf]] - AbDPS + ketobutanoic acid<br /> | |||
**[[3u8g]] - AbDPS + oxalic acid<br /> | |||
**[[3uqn]] - AbDPS + oxamic acid<br /> | |||
**[[3rk8]], [[3tak]] - AbDPS + pyruvate<br /> | |||
**[[3puo]] - PaDPS + Lys<br /> | |||
**[[3s8h]] - PaDPS + hydroxypropanoic acid<br /> | |||
**[[4fha]] - SpDPS + Lys<br /> | |||
**[[2vc6]] – SmDPS + pyruvate <br /> | |||
**[[4ly8]], [[4mlj]], [[7kwn]], [[7l4b]] - CjDPS + pyruvate <br /> | |||
**[[6tzu]], [[6u01]], [[7kel]], [[7kg5]], [[7klq]], [[7klt]], [[7kn2]], [[7ko1]], [[7koc]], [[7kr8]], [[7ku6]], [[7kx1]], [[7lbd]] - CjDPS (mutant) + pyruvate <br /> | |||
**[[7m06]] - CjDPS (mutant) + Lys derivative<br /> | |||
**[[5f1u]], [[5f1v]] - CjDPS + inhibitor<br /> | |||
**[[4mlr]] - CjDPS + pyruvate + Lys<br /> | |||
**[[3di1]] - SaDPS + pyruvate<br /> | |||
**[[1yxd]], [[2ats]] – EcDPS + S-Lys<br /> | |||
**[[3c0j]] - EcDPS + hydroxypyruvate<br /> | |||
**[[3du0]], [[7jzf]] - EcDPS + pyruvate<br /> | |||
**[[3i7s]] - EcDPS (mutant) + pyruvate<br /> | |||
**[[4eou]] - EcDPS + succinic semialdehyde<br /> | |||
**[[6nva]] - EcDPS + propionate<br /> | |||
**[[5t25]], [[5t26]] - EcDPS + inhibitor<br /> | |||
**[[3hij]] - BaDPS + pyruvate<br /> | |||
**[[6xgs]] - DPS + citrate – ''Brucella suis''<br /> | |||
**[[7lvl]] - ClDPS + S-Lys <br /> | |||
**[[4nq1]] - DPS + pyruvate – ''Legionella pneumophila''<br /> | |||
**[[6ue0]] - DPS + pyruvate – ''Klebsiella pneumoniae''<br /> | |||
**[[5j5d]] - MtDPS + ketopimelic acid<br /> | |||
</ | *DHDPS ternary complexes | ||
**[[4pfm]] - SbDPS + Lys + pyruvate<br /> | |||
**[[7kg2]] - CjDPS (mutant) + pyruvate + His<br /> | |||
**[[7kxh]], [[7lcf]], [[7kwp]] – CjDPS + pyruvate + Lys derivative<br /> | |||
**[[7kg9]], [[7kh4]], [[7kk1]], [[7kkd]], [[7kkg]], [[7kkt]], [[7kls]], [[7kly]], [[7km0]], [[7km1]], [[7kn9]], [[7knz]], [[7kpc]], [[7kpe]], [[7ko3]], [[7kr7]], [[7kto]], [[7kuz]], [[7kwf]], [[7kz2]] - CjDPS (mutant) + pyruvate + Lys derivative<br /> | |||
**[[4m19]], [[4mlr]] - CjDPS + pyruvate + Lys<br /> | |||
**[[7kxg]] - CjDPS + pyruvate + His<br /> | |||
**[[7jzc]], [[7jzg]] - EcDPS (mutant) + Lys + pyruvate<br /> | |||
**[[7jz9]], [[7jzb]] - EcDPS (mutant) + pyruvate + succinic semialdehyde <br /> | |||
**[[7jza]] - EcDPS (mutant) + pyruvate + Lys + succinic semialdehyde <br /> | |||
**[[7mjf]] - ClDPS + pyruvate + succinic semialdehyde <br /> | |||
}} | |||
==Reference== | ==Reference== | ||
Crystal structure of YagE, a putative DHDPS-like protein from Escherichia coli K12., Manicka S, Peleg Y, Unger T, Albeck S, Dym O, Greenblatt HM, Bourenkov G, Lamzin V, Krishnaswamy S, Sussman JL, Proteins. 2008 Jun;71(4):2102-8. PMID:[http://www.ncbi.nlm.nih.gov/pubmed/18361457 18361457 ] | Crystal structure of YagE, a putative DHDPS-like protein from Escherichia coli K12., Manicka S, Peleg Y, Unger T, Albeck S, Dym O, Greenblatt HM, Bourenkov G, Lamzin V, Krishnaswamy S, Sussman JL, Proteins. 2008 Jun;71(4):2102-8. PMID:[http://www.ncbi.nlm.nih.gov/pubmed/18361457 18361457 ] | ||
Latest revision as of 13:19, 22 December 2022
CRYSTAL STRUCTURE OF YAGE, A PROPHAGE PROTEIN BELONGING TO THE DIHYDRODIPICOLINIC ACID SYNTHASE FAMILY FROM E. COLI K12
One approach of the incorporation of genetic material from different species into bacterial genomes is infection by a bacteriophage. The yagE gene is located in the E. coli K12 genome and it is part of prophage CP4-6 encoding a 33-kDa putative dihydrodipicolinate synthase or 4-hydroxy-tetrahydrodihydrodipicolinate synthase (DHDPS)-like protein (UniProtKB/Swiss-prot: P75682). This DHDPS-like domain is a member of the N-acetyl neuraminate lyase (NAL) subfamily comprises an 8-fold (TIM barrel) with a small C-terminal α-helical domain (Inter pro: IPR005263). Many enzymes possess this DHDPS domain (e.g. HBPHA, KDGA, NAL, DOGDH, and DHDPS). All these enzymes contain an 8-fold α/β barrel and a small C-terminal α-helical region. between orthorhombic (2v8z) and monoclinic (2v9d) crystal forms of YagE revealed striking similarity. Cartoon representation of YagE showing the 2-fold symmetry axes between monomers. Chains A, B, C, and D are colored cyan, lime, magenta, and yellow, respectively. of YagE (cyan) with (1nal, yellow), (1w3i, magenta), (1xky, lime), and (2ats, blue) monomers. Although YagE possesses only 23, 24, and 27% sequence identity with EcNAL, SsKDGA, EcDHDPS, respectively, their monomeric structures are all very similar. Superposition of active site residues of YagE with (magenta, 1w3i), (blue, 2ats), and (yellow, 1nal), residues are labeled according to the corresponding PDB structures. Members of the NAL protein subfamily have very similar active sites and a single amino acid substitution can significantly change their function. For example, NAL (1nal) gets DHDPS activity by substitution of a (orange) to (blue) at position 142. The possible active site region of YagE demonstrates closest sequence similarity to the active site of KDG aldolase of SsKDGA (1w3i) and NAL of EcNAL (1nal) (yellow), suggesting that this protein can perform either of these functions. Although the active site of EcDHDPS (1xky) and BaDHDPS (2ats) shows similarities, the important residue that differentiates between NAL and DHDPS, namely Leu142 (in 1nal), is also present in YagE (labeled cyan) at that particular position suggesting that YagE performs a NAL-related function rather than DHDPS-related one. In conclusion, the high-resolution X-ray structure of YagE provides a clue that it probably belongs to the NAL subfamily of proteins. Although the exact molecular function of YagE is still unknown, its structure provides a handle for understanding its molecular function based on knowledge about conserved residues of the putative active site. |
| ||||||||||
3D structures of dihydrodipicolinate synthase3D structures of dihydrodipicolinate synthase
Updated on 22-December-2022
ReferenceReference
Crystal structure of YagE, a putative DHDPS-like protein from Escherichia coli K12., Manicka S, Peleg Y, Unger T, Albeck S, Dym O, Greenblatt HM, Bourenkov G, Lamzin V, Krishnaswamy S, Sussman JL, Proteins. 2008 Jun;71(4):2102-8. PMID:18361457
- Created with the participation of Jaime Prilusky and Michal Harel.
Proteopedia Page Contributors and Editors (what is this?)Proteopedia Page Contributors and Editors (what is this?)
Alexander Berchansky, Michal Harel- Topic Page
- Escherichia coli
- Single protein
- Albeck, S.
- Bourenkov, G.
- Dym, O.
- Greenblatt, H M.
- Krishnaswamy, S.
- Lamzin, V.
- Manicka, S.
- Peleg, Y.
- Sussman, J L.
- Unger, T.
- ISPC, Israel Structural Proteomics Center.
- ISPC
- Israel Structural Proteomics Center
- Dhdp
- Dihydrodipicolinic acid synthase
- Lyase
- N-acetyl neuraminate lyase
- Nal
- Prophage