Proteopedia:Featured SEL/8: Difference between revisions
Jump to navigation
Jump to search
m Protected "Proteopedia:Featured SEL/8" [edit=sysop:move=sysop] |
Eric Martz (talk | contribs) No edit summary |
||
| Line 8: | Line 8: | ||
<tr><td><div class='scrolling '>'''BREAKTHROUGH in protein structure prediction!'''<br> | <tr><td><div class='scrolling '>'''BREAKTHROUGH in protein structure prediction!'''<br> | ||
''by Eric Martz''<br> | ''by Eric Martz''<br> | ||
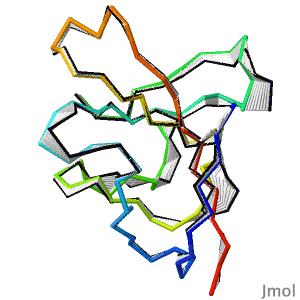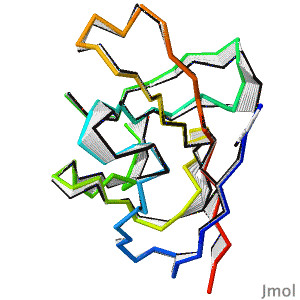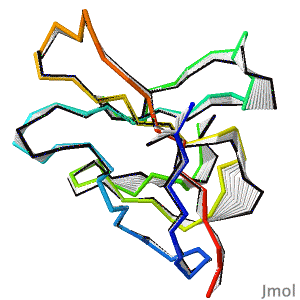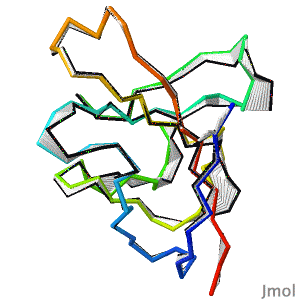
After decades of slow progress by many groups, in 2020, AlphaFold2 proved able to accurately predict the detailed structures of two-thirds of single protein domains from their amino acid sequences. Pictured is AlphaFold2's prediction for the ORF8 protein of SARS-CoV-2 (black), compared with a subsequently published X-ray crystallographic structure (colors). ORF8 contributes to virulence in COVID-19. | After decades of slow progress by many groups, in 2020, AlphaFold2 proved able to accurately predict the detailed structures of two-thirds of single protein domains from their amino acid sequences. Pictured is AlphaFold2's prediction for the ORF8 protein of SARS-CoV-2 (black), compared with a subsequently published X-ray crystallographic structure (colors). ORF8 contributes to virulence in COVID-19.<br> | ||
>>> [[Theoretical_models#2020:_CASP_14|Visit this page]] >>> | >>> [[Theoretical_models#2020:_CASP_14|Visit this page]] >>> | ||
</div> | </div> | ||
Latest revision as of 00:08, 9 November 2022
 |
BREAKTHROUGH in protein structure prediction!
by Eric Martz |