User:Ke Xiao/Geobacter pilus models: Difference between revisions
Eric Martz (talk | contribs) No edit summary |
Eric Martz (talk | contribs) No edit summary |
||
| (14 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
Interactive 3D | [[Interactive_3D_Complement_in_Proteopedia|Interactive 3D Complement in Proteopedia]]<br> | ||
{{Clear}}<br> | |||
<span style="border: 2px solid #a1a1a1; border-radius: 6px;padding:35px 50px 20px 30px;background: linear-gradient(#b0d0ff, #ffffff);"><span style="font-size:250%;">Scientific Reports</span> | |||
<span style="padding: 0px 0px 0px 100px;color:#808080;font-size:120%;">an online, open access journal: [http://www.nature.com/srep nature.com/srep]</span></span> | |||
{{Clear}}<br> | |||
<span style="font-size:160%;line-height:130%;"><b>Low energy atomic models suggesting a pilus structure that could account for electrical conductivity of ''Geobacter sulfurreducens'' pili.</b></span> | |||
<br> | |||
<span style="font-size:120%; line-height:160%;">Ke '''[[User:Ke_Xiao|Xiao]]''', Nikhil S. '''[[User:Nikhil_Malvankar|Malvankar]]''', Chuanjun '''[https://www.researchgate.net/profile/Chuanjun_Shu Shu]''', Eric '''[[User:Eric Martz|Martz]]''', Derek R. '''[http://www.micro.umass.edu/faculty-and-research/derek-lovley Lovley]''', and Xiao '''[http://www.lmbe.seu.edu.cn/chenyuan/xsun/mainpage.htm Sun]'''. | |||
<br> | |||
''Scientific Reports'' '''6''':23385, March 2016: [http://www.nature.com/articles/srep23385 nature.com/articles/srep23385]. ([http://dx.doi.org/10.1038/srep23385 DOI: 10.1038/srep23385]) | |||
<!--[http://mbio.asm.org/content/6/2/e00084-15.abstract mBio 6(2):e00084-15] (2015). ([http://dx.doi.org/10.1128/mBio.00084-15 doi:10.1128/mBio.00084-15])--> | |||
</span> | |||
__NOTOC__ | |||
<table class="wikitable" style="background-color:#ffffa0;"><tr><td> | |||
CAVEAT: The theoretical model described here was published in 2016. In 2019, [[cryo-EM]] structures of highly conductive ''Geobacter'' nanowires were found to be composed of the cytochrome OmcS<ref name="wang-gu">PMID:30951668</ref><ref name="filman">PMID:31925024</ref>. This called into question whether highly conductive pilus nanowires composed of the protein pilA exist. | |||
</td></tr></table> | |||
==Molecular Tour== <!--####################################--> | |||
<StructureSection size='[300,600]' side='right' caption='Drag with mouse to rotate. Zoom with mouse wheel, or shift+drag.' scene='69/699340/Arc-1_no_hydrogen/1'> | |||
__TOC__ | |||
<center> | |||
<span style="border: 2px solid #ffff00; border-radius: 5px;padding:3px 8px 3px 8px;background: #ffffa0;"> | |||
Click the ''green links'' below to change the molecular scene. | |||
</span> | |||
</center> | |||
===Pilus Model=== | |||
The theoretical ''Geobacter sulfurreducens'' pilus model shown here, called ''ARC-1''<ref name="xiao1">PMID: 27001169</ref> (<scene name='69/699340/Arc-1_no_hydrogen/1'>restore initial scene</scene>), is representative of a cluster of 50 low-energy models with an arrangement of aromatic rings consistent with X-ray diffraction data<ref name="malvankar2015">PMID: 25736881</ref>. Unlike the docking of crystallographic models into electron density from cryo-electron microscopy<ref>PMID: 16949362</ref><ref>PMID: 12769840</ref>, these models have chemically realistic interactions between subunit chains. Energy minimization produced subunit interactions with shape complementarity, non-covalent bonds, and very few atomic clashes<ref>The chemical realism of these theoretical energy-minimized models contrasts with models where empirical monomer structures are docked into cryo-electron microscopic electron density maps. Those are unrealistic in details of subunit interactions, lacking shape complementarity and having many atomic clashes. See "Initial Model Outputs" in [http://www.nature.com/articles/srep23385 the publication] for details.</ref>. | |||
===Monomer Chains=== | |||
This ARC-1 model contains 21 chains of pilA<ref>Each chain contains the 61 C-terminal amino acids of [http://www.uniprot.org/uniprot/Q74D23 UniProt Q74D23].</ref> of ''Geobacter sulfurreducens''. The 21 chains were restrained to have <scene name='69/699340/Arc-1_no_hydrogen/2'>identical conformations</scene>. Amino acids 3-50 are <scene name='69/699340/Arc-1_no_hydrogen/3'>alpha-helical, with a slight bend at Pro22</scene>. Modeling was initiated with model 1 of NMR ensemble [[2m7g]] of ''Geobacter sulfurreducens''. The monomers in ARC-1 are only slightly different from the initial conformation: | |||
* <scene name='69/699340/2m7g_mdl1_vs_arc1_chaink/1'>Compare main chain traces</scene>: '''<font color="#006000">2m7g model 1</font>, <font color="magenta">ARC-1 chain K</font>'''. | |||
* <scene name='69/699340/2m7g_mdl1_vs_arc1_chaink/2'>Compare sidechains</scene>. | |||
===Stacked Aromatic Rings=== | |||
The ARC-1 model (and others of its cluster) are the first chemically realistic pilus models that can account for the electrical conductivity of these pili in terms of a core of stacked aromatic rings. These models are consistent with multiple lines of experimental evidence including X-ray diffraction suggesting stacked aromatics<ref name="xiao1" /><ref name="malvankar2015" />. | |||
Each pilA chain contains <scene name='69/699340/Arc-1_no_hydrogen/9'>six aromatic amino acids</scene>. In the pilus assembly, <scene name='69/699340/Arc-1_no_hydrogen/10'>half of the aromatic rings form a helical chain in the core, while the other half are near the surface</scene>. | |||
The aromatic <scene name='69/699340/Arc-1_no_hydrogen/11'>rings in the core are closely packed</scene>. The core aromatic rings are <scene name='69/699340/Arc-1_no_hydrogen/13'>Phe1, Phe24 and Tyr27</scene>. | |||
===Salt Bridges=== | |||
80% of the 50 lowest-energy models have a [[Salt bridges|salt bridge]] between Arg41 and Asp39 in different chains. | |||
* <scene name='69/699340/Arc-1_no_hydrogen/14'>Salt bridges stabilize the pilus</scene> (Arg41:Asp39, sidechain nitrogens and oxygens). | |||
* <scene name='69/699340/Arc-1_no_hydrogen/15'>Single salt bridge between chains J and K</scene>. | |||
===Monomers Per Turn=== | |||
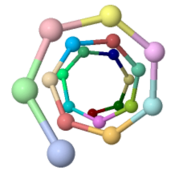
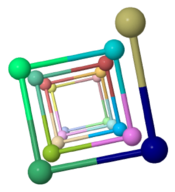
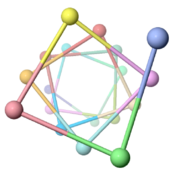
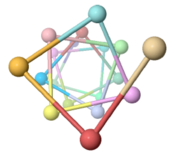
The ARC-1 model has 6.4 monomer chains per turn (56.0 degrees rotation between monomers). This is more chains/turn than some previous type IV pilus models. Among the 50 models with lowest energy in the cluster including ARC-1, chains/turn ranged from 5.0 to 7.6. | |||
To visualize chains/turn, we show <scene name='69/699340/Arc-1_no_hydrogen/18'>just one atom per protein monomer chain</scene> (alpha carbon of Phe51). Then these chain-marking-atoms are <scene name='69/699340/Arc-1_no_hydrogen/20'>connected with with rods</scene>, and the resulting helix is viewed from one end. When counting the chains/turn, bear in mind that the first and last (to complete one turn) count as 1/2 chain each. | |||
<table cellpadding="4" border="1"> | |||
<tr><td> | |||
Pilus | |||
</td><td> | |||
Chains/Turn (Angle) | |||
</td><td> | |||
Image | |||
</td></tr><tr><td> | |||
''Geobacter sulfurreducens'' (type IVa, 61 amino acids, theoretical model ARC-1, 2016)<ref name="xiao1" /> | |||
</td><td> | |||
<span style="font-size:170%;">'''6.4''' (56.0°)</span> | |||
</td><td> | |||
[[Image:CPT-Gs-Xiao-end.png|175px]] | |||
</td></tr><tr><td> | |||
''Klebsielle oxytoca'' (type IVa, 137 amino acids [http://www.uniprot.org/uniprot/A0A0E0WUQ8 A0A0E0WUQ8], theoretical model, 2010)<ref>PMID: 21115127</ref> | |||
</td><td> | |||
<span style="font-size:170%;">'''4.3''' (84.7°)</span> | |||
</td><td> | |||
</td></tr><tr><td> | |||
''Pseudomonas aeruginosa'' (type IVa, 150 amino acids, fiber diffraction, 2004)<ref>PMID: 15100690</ref> | |||
</td><td> | |||
<span style="font-size:170%;">'''4.0''' (90°)</span> | |||
</td><td> | |||
[[Image:CPT-Pa-end.png|175px]] | |||
</td></tr><tr><td> | |||
''Vibrio cholerae'' (type '''IVb''', 198 amino acids, cryo-EM, 2012)<ref>PMID: 22361030</ref> | |||
</td><td> | |||
<span style="font-size:170%;">'''3.7''' (96.8°)</span> | |||
</td><td> | |||
[[Image:CPT-Vc2012-end.png|175px]] | |||
</td></tr><tr><td> | |||
''Neisseria gonorrhoeae'' (type IVa, 165 amino acids, cryo-EM [[2hil]], 2006)<ref>PMID: 16949362</ref> | |||
</td><td> | |||
<span style="font-size:170%;">'''3.6''' (100.8°)</span> | |||
</td><td> | |||
[[Image:CPT-Ng-2hil-end.png|175px]] | |||
</td></tr></table> | |||
</StructureSection> | |||
<!--####################################--> | |||
<br> | |||
{{Theoretical_model}} | |||
<hr><br> | |||
==Download== | |||
===Pilus Model=== | |||
*Click to download [http://proteopedia.org/wiki/images/8/8e/Geobacter_sulfurreducens_pilus_model_ARC-1.pdb Geobacter sulfurreducens pilus model ARC-1] | |||
*[http://bioinformatics.org/firstglance/fgij/fg.htm?mol=http://proteopedia.org/wiki/images/8/8e/Geobacter_sulfurreducens_pilus_model_ARC-1.pdb Explore pilus model in FirstGlance in Jmol]. | |||
===Animations for Powerpoint=== | |||
{| class="wikitable" | |||
|- | |||
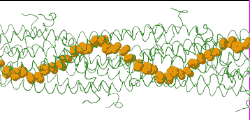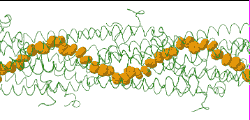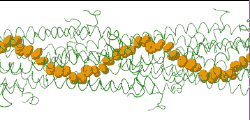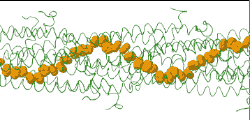
| [[Image:Model128-1-24-27-orange-greentrace-animation-250px.gif]] | |||
| Low resolution, ARC-1 model (<font color="green">'''smoothed green backbone traces'''</font>) with <font color="#d08000">'''aromatic rings of residues Phe1, Phe24, and Tyr27 in orange'''</font>. [http://bioinformatics.org/molvis/gifs/model128-1-24-27-orange-greentrace-animation.gif DOWNLOAD HIGH RESOLUTION ANIMATION] (19 MB). | |||
|- | |||
| [[Image:Model128-1-24-27-path-solid-animation-250px.gif]] | |||
| Low resolution, ARC-1 model with spacefilling (van der Waals) atoms, each chain a different color. [http://bioinformatics.org/molvis/gifs/model128-1-24-27-path-solid-animation.gif DOWNLOAD HIGH RESOLUTION ANIMATION] (29 MB). | |||
|- | |||
| [[Image:Model128-1-24-27-path-translucent-animation-250px.gif]] | |||
| Low resolution, ARC-1 model with translucent spacefilling atoms. Aromatic rings of Phe1, Phe24, and Tyr27 are opaque. Each chain is a different color. [http://bioinformatics.org/molvis/gifs/model128-1-24-27-path-translucent-animation.gif DOWNLOAD HIGH RESOLUTION ANIMATION] (28 MB). | |||
|} | |||
==See Also== | |||
* [http://geobacter.org Geobacter.Org] | |||
==Notes & References== | |||
<references /> | |||
Latest revision as of 01:26, 9 August 2021
Interactive 3D Complement in Proteopedia
Scientific Reports an online, open access journal: nature.com/srep
Low energy atomic models suggesting a pilus structure that could account for electrical conductivity of Geobacter sulfurreducens pili.
Ke Xiao, Nikhil S. Malvankar, Chuanjun Shu, Eric Martz, Derek R. Lovley, and Xiao Sun.
Scientific Reports 6:23385, March 2016: nature.com/articles/srep23385. (DOI: 10.1038/srep23385)
|
CAVEAT: The theoretical model described here was published in 2016. In 2019, cryo-EM structures of highly conductive Geobacter nanowires were found to be composed of the cytochrome OmcS[1][2]. This called into question whether highly conductive pilus nanowires composed of the protein pilA exist. |
Molecular TourMolecular Tour
Click the green links below to change the molecular scene. Pilus ModelThe theoretical Geobacter sulfurreducens pilus model shown here, called ARC-1[3] (), is representative of a cluster of 50 low-energy models with an arrangement of aromatic rings consistent with X-ray diffraction data[4]. Unlike the docking of crystallographic models into electron density from cryo-electron microscopy[5][6], these models have chemically realistic interactions between subunit chains. Energy minimization produced subunit interactions with shape complementarity, non-covalent bonds, and very few atomic clashes[7]. Monomer ChainsThis ARC-1 model contains 21 chains of pilA[8] of Geobacter sulfurreducens. The 21 chains were restrained to have . Amino acids 3-50 are . Modeling was initiated with model 1 of NMR ensemble 2m7g of Geobacter sulfurreducens. The monomers in ARC-1 are only slightly different from the initial conformation:
Stacked Aromatic RingsThe ARC-1 model (and others of its cluster) are the first chemically realistic pilus models that can account for the electrical conductivity of these pili in terms of a core of stacked aromatic rings. These models are consistent with multiple lines of experimental evidence including X-ray diffraction suggesting stacked aromatics[3][4]. Each pilA chain contains . In the pilus assembly, . The aromatic . The core aromatic rings are . Salt Bridges80% of the 50 lowest-energy models have a salt bridge between Arg41 and Asp39 in different chains.
Monomers Per TurnThe ARC-1 model has 6.4 monomer chains per turn (56.0 degrees rotation between monomers). This is more chains/turn than some previous type IV pilus models. Among the 50 models with lowest energy in the cluster including ARC-1, chains/turn ranged from 5.0 to 7.6. To visualize chains/turn, we show (alpha carbon of Phe51). Then these chain-marking-atoms are , and the resulting helix is viewed from one end. When counting the chains/turn, bear in mind that the first and last (to complete one turn) count as 1/2 chain each.
|
| ||||||||||||||||||||||||||||
DownloadDownload
Pilus ModelPilus Model
- Click to download Geobacter sulfurreducens pilus model ARC-1
- Explore pilus model in FirstGlance in Jmol.
Animations for PowerpointAnimations for Powerpoint
|
Low resolution, ARC-1 model (smoothed green backbone traces) with aromatic rings of residues Phe1, Phe24, and Tyr27 in orange. DOWNLOAD HIGH RESOLUTION ANIMATION (19 MB). |

|
Low resolution, ARC-1 model with spacefilling (van der Waals) atoms, each chain a different color. DOWNLOAD HIGH RESOLUTION ANIMATION (29 MB). |

|
Low resolution, ARC-1 model with translucent spacefilling atoms. Aromatic rings of Phe1, Phe24, and Tyr27 are opaque. Each chain is a different color. DOWNLOAD HIGH RESOLUTION ANIMATION (28 MB). |
See AlsoSee Also
Notes & ReferencesNotes & References
- ↑ Wang F, Gu Y, O'Brien JP, Yi SM, Yalcin SE, Srikanth V, Shen C, Vu D, Ing NL, Hochbaum AI, Egelman EH, Malvankar NS. Structure of Microbial Nanowires Reveals Stacked Hemes that Transport Electrons over Micrometers. Cell. 2019 Apr 4;177(2):361-369.e10. doi: 10.1016/j.cell.2019.03.029. PMID:30951668 doi:http://dx.doi.org/10.1016/j.cell.2019.03.029
- ↑ Filman DJ, Marino SF, Ward JE, Yang L, Mester Z, Bullitt E, Lovley DR, Strauss M. Cryo-EM reveals the structural basis of long-range electron transport in a cytochrome-based bacterial nanowire. Commun Biol. 2019 Jun 19;2(1):219. doi: 10.1038/s42003-019-0448-9. PMID:31925024 doi:http://dx.doi.org/10.1038/s42003-019-0448-9
- ↑ 3.0 3.1 3.2 Xiao K, Malvankar NS, Shu C, Martz E, Lovley DR, Sun X. Low Energy Atomic Models Suggesting a Pilus Structure that could Account for Electrical Conductivity of Geobacter sulfurreducens Pili. Sci Rep. 2016 Mar 22;6:23385. doi: 10.1038/srep23385. PMID:27001169 doi:http://dx.doi.org/10.1038/srep23385
- ↑ 4.0 4.1 Malvankar NS, Vargas M, Nevin K, Tremblay PL, Evans-Lutterodt K, Nykypanchuk D, Martz E, Tuominen MT, Lovley DR. Structural basis for metallic-like conductivity in microbial nanowires. MBio. 2015 Mar 3;6(2):e00084. doi: 10.1128/mBio.00084-15. PMID:25736881 doi:http://dx.doi.org/10.1128/mBio.00084-15
- ↑ Craig L, Volkmann N, Arvai AS, Pique ME, Yeager M, Egelman EH, Tainer JA. Type IV pilus structure by cryo-electron microscopy and crystallography: implications for pilus assembly and functions. Mol Cell. 2006 Sep 1;23(5):651-62. PMID:16949362 doi:10.1016/j.molcel.2006.07.004
- ↑ Craig L, Taylor RK, Pique ME, Adair BD, Arvai AS, Singh M, Lloyd SJ, Shin DS, Getzoff ED, Yeager M, Forest KT, Tainer JA. Type IV pilin structure and assembly: X-ray and EM analyses of Vibrio cholerae toxin-coregulated pilus and Pseudomonas aeruginosa PAK pilin. Mol Cell. 2003 May;11(5):1139-50. PMID:12769840
- ↑ The chemical realism of these theoretical energy-minimized models contrasts with models where empirical monomer structures are docked into cryo-electron microscopic electron density maps. Those are unrealistic in details of subunit interactions, lacking shape complementarity and having many atomic clashes. See "Initial Model Outputs" in the publication for details.
- ↑ Each chain contains the 61 C-terminal amino acids of UniProt Q74D23.
- ↑ Campos M, Francetic O, Nilges M. Modeling pilus structures from sparse data. J Struct Biol. 2011 Mar;173(3):436-44. doi: 10.1016/j.jsb.2010.11.015. Epub 2010 , Nov 27. PMID:21115127 doi:http://dx.doi.org/10.1016/j.jsb.2010.11.015
- ↑ Craig L, Pique ME, Tainer JA. Type IV pilus structure and bacterial pathogenicity. Nat Rev Microbiol. 2004 May;2(5):363-78. PMID:15100690 doi:http://dx.doi.org/10.1038/nrmicro885
- ↑ Li J, Egelman EH, Craig L. Structure of the Vibrio cholerae Type IVb Pilus and stability comparison with the Neisseria gonorrhoeae type IVa pilus. J Mol Biol. 2012 Apr 20;418(1-2):47-64. doi: 10.1016/j.jmb.2012.02.017. Epub 2012, Feb 21. PMID:22361030 doi:http://dx.doi.org/10.1016/j.jmb.2012.02.017
- ↑ Craig L, Volkmann N, Arvai AS, Pique ME, Yeager M, Egelman EH, Tainer JA. Type IV pilus structure by cryo-electron microscopy and crystallography: implications for pilus assembly and functions. Mol Cell. 2006 Sep 1;23(5):651-62. PMID:16949362 doi:10.1016/j.molcel.2006.07.004