Myosin: Difference between revisions
Michal Harel (talk | contribs) No edit summary |
Michal Harel (talk | contribs) No edit summary |
||
| (3 intermediate revisions by the same user not shown) | |||
| Line 10: | Line 10: | ||
*'''Myosin X''' (MX) is a downstream effector of PI(3)K during phagocytosis<ref>PMID:12055636</ref>. | *'''Myosin X''' (MX) is a downstream effector of PI(3)K during phagocytosis<ref>PMID:12055636</ref>. | ||
*'''Myosin XI''' (MXI) links the nuclear membrane to the cytoskeleton<ref>PMID:23973298</ref>. | *'''Myosin XI''' (MXI) links the nuclear membrane to the cytoskeleton<ref>PMID:23973298</ref>. | ||
See also [[Myosin (hebrew)]]. | |||
==Crystallization and X-ray diffraction== | ==Crystallization and X-ray diffraction== | ||
| Line 39: | Line 41: | ||
==Disease== | ==Disease== | ||
Mutations in MIIA cause early onset myopathy<ref>PMID:20418530</ref>. Mutations in MVIIA cause Usher syndrome<ref>PMID:7870171</ref>. | Mutations in MIIA cause early onset myopathy<ref>PMID:20418530</ref>. Mutations in MVIIA cause Usher syndrome<ref>PMID:7870171</ref>. | ||
== 3D Structures of Myosin == | == 3D Structures of Myosin == | ||
[[Myosin 3D Structures]] | |||
</StructureSection> | |||
== References == | == References == | ||
<references/> | <references/> | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Latest revision as of 12:09, 16 February 2021
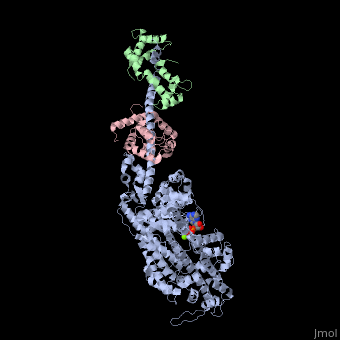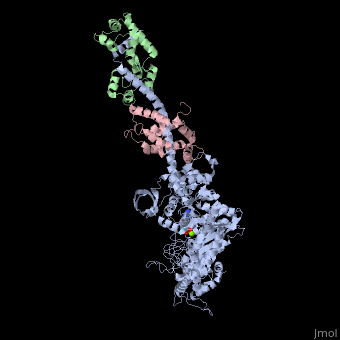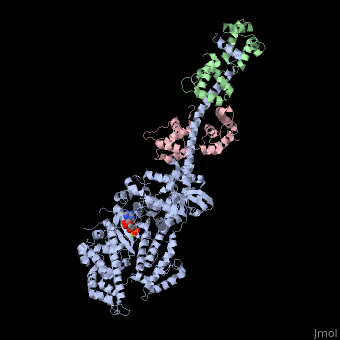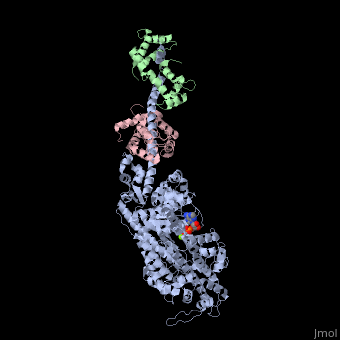
IntroductionMyosin is one of three major classes of molecular motor proteins: myosin, dynein, and kinesin. As the most abundant of these proteins myosin plays a structural and enzymatic role in muscle contraction and intracellular motility. Myosin was first discovered in muscle in the 19th century. [1] Myosin is a superfamily of proteins which bind actin, hydrolyze ATP and transduce force. Thus most are located in muscle cells. Composed of head, neck and tail domains. Head domain binds the actin and moves along it. The neck is a linker and binds the light chains which have a regulatory function. The tail interacts with cargo molecules (CBD)m. There are 18 classes of myosin. Unconventional myosin are thought not to form filaments[2].
See also Myosin (hebrew). Crystallization and X-ray diffractionMyosin is found in abundance, therefore it can be prepared in gram quantities. [9] For nearly 30 years the myosin head was resistant to crystallization, yet by 1993 researchers discovered a mechanism to obtain x-ray quality crystals. The process modified the protein by reductive methylation. X-ray data was used to determine the tertiary structure of the protein. [9] Structure Myosin has a molecular size of approximately 520 kilodaltons with a total of six subunits. It has two 220 kD heavy chains which make the majority of the overall structure and two pairs of light chains which vary in size.[9] The molecule is asymmetric, having a long tail and two globular heads. Each heavy chains composes the bulk of one of the globular heads. Sub-fragment-1(S1) also termed the myosin head consists of ATP, actin, and two light chain binding sites. Each globular head has a heavy chain and two light chains for a combined molecular size of about 130 kD. [9] The myosin head is asymmetrical with a length of 165 Angstroms and 65 Angstroms in width, with a total thickness of about 40 Angstroms. [9] About 48% of the amino acid residues in the myosin head are dominated by α helices. At the carboxyl terminus one long α helix of about 85 Angstroms extends in a left-handed coil. This particular helix forms the light chain binding region of the globular domain [9] The amino terminus of each heavy chain has a large globular domain containing the site of ATP hydrolysis. FunctionMolecules of myosin aggregate in muscle cells to form thick filaments. [10] The rodlike structure of these thick filaments act as the core in the muscle contractile unit. The aggregation of several hundred myosin forms a bipolar structure which stacks in regular arrays. Muscles consist of another protein called actin. Actin forms the thin filament in muscle fibers. Myosin and actin interact through weak bonds. Without ATP bound, the myosin head binds tightly to actin. With ATP bound, myosin releases the actin subunit and interacts with another subunit further down the thin filament. This process continues in cycle, producing movement. Interaction of myosin and actin is regulated by two other proteins, tropomyosin and troponin. [10] The cycle of myosin-actin interaction is outlined as follows: [10] 1. ATP binds to myosin and a binding site opens on myosin head to disrupt the actin-myosin interaction, actin is released. ATP is hydrolyzed 2. a conformational change moving the protein to a "high-energy" state causes the myosin head to change orientation moving it to bind with the actin subunit closer the a region called the Z disk than the previous actin subunit 3. the binding site is closed, strengthening the myosin-actin binding 4. a quickly follows and the myosin head undergoes an additional conformational change bringing it back to the resting state in which it began Click the link to access DNAtube video "A Moving Myosin Motor Protein" http://www.dnatube.com/video/389/A-Moving-Myosin-Motor-Protein-myosin-actin-interaction DiseaseMutations in MIIA cause early onset myopathy[11]. Mutations in MVIIA cause Usher syndrome[12]. 3D Structures of Myosin
|
| ||||||||||
ReferencesReferences
- ↑ Spudich JA, Finer J, Simmons B, Ruppel K, Patterson B, Uyeda T. Myosin structure and function. Cold Spring Harb Symp Quant Biol. 1995;60:783-91. PMID:8824453
- ↑ Kalhammer G, Bahler M. Unconventional myosins. Essays Biochem. 2000;35:33-42. PMID:12471888
- ↑ Matsumura F. Regulation of myosin II during cytokinesis in higher eukaryotes. Trends Cell Biol. 2005 Jul;15(7):371-7. PMID:15935670 doi:http://dx.doi.org/10.1016/j.tcb.2005.05.004
- ↑ Mehta AD, Rock RS, Rief M, Spudich JA, Mooseker MS, Cheney RE. Myosin-V is a processive actin-based motor. Nature. 1999 Aug 5;400(6744):590-3. PMID:10448864 doi:http://dx.doi.org/10.1038/23072
- ↑ Buss F, Spudich G, Kendrick-Jones J. Myosin VI: cellular functions and motor properties. Annu Rev Cell Dev Biol. 2004;20:649-76. PMID:15473855 doi:http://dx.doi.org/10.1146/annurev.cellbio.20.012103.094243
- ↑ Hasson T, Skowron JF, Gilbert DJ, Avraham KB, Perry WL, Bement WM, Anderson BL, Sherr EH, Chen ZY, Greene LA, Ward DC, Corey DP, Mooseker MS, Copeland NG, Jenkins NA. Mapping of unconventional myosins in mouse and human. Genomics. 1996 Sep 15;36(3):431-9. PMID:8884266 doi:http://dx.doi.org/10.1006/geno.1996.0488
- ↑ Cox D, Berg JS, Cammer M, Chinegwundoh JO, Dale BM, Cheney RE, Greenberg S. Myosin X is a downstream effector of PI(3)K during phagocytosis. Nat Cell Biol. 2002 Jul;4(7):469-77. PMID:12055636 doi:http://dx.doi.org/10.1038/ncb805
- ↑ Tamura K, Iwabuchi K, Fukao Y, Kondo M, Okamoto K, Ueda H, Nishimura M, Hara-Nishimura I. Myosin XI-i links the nuclear membrane to the cytoskeleton to control nuclear movement and shape in Arabidopsis. Curr Biol. 2013 Sep 23;23(18):1776-81. doi: 10.1016/j.cub.2013.07.035. Epub 2013 , Aug 22. PMID:23973298 doi:http://dx.doi.org/10.1016/j.cub.2013.07.035
- ↑ 9.0 9.1 9.2 9.3 9.4 9.5 Rayment I, Rypniewski WR, Schmidt-Base K, Smith R, Tomchick DR, Benning MM, Winkelmann DA, Wesenberg G, Holden HM. Three-dimensional structure of myosin subfragment-1: a molecular motor. Science. 1993 Jul 2;261(5117):50-8. PMID:8316857
- ↑ 10.0 10.1 10.2 Nelson, D. and Cox, M.(2005). Lehninger Principles of Biochemistry. 4th ed. p.1119.
- ↑ Tajsharghi H, Hilton-Jones D, Raheem O, Saukkonen AM, Oldfors A, Udd B. Human disease caused by loss of fast IIa myosin heavy chain due to recessive MYH2 mutations. Brain. 2010 May;133(Pt 5):1451-9. doi: 10.1093/brain/awq083. PMID:20418530 doi:http://dx.doi.org/10.1093/brain/awq083
- ↑ Weil D, Blanchard S, Kaplan J, Guilford P, Gibson F, Walsh J, Mburu P, Varela A, Levilliers J, Weston MD, et al.. Defective myosin VIIA gene responsible for Usher syndrome type 1B. Nature. 1995 Mar 2;374(6517):60-1. PMID:7870171 doi:http://dx.doi.org/10.1038/374060a0