Tryptophan RNA-binding attenuation protein: Difference between revisions
Jump to navigation
Jump to search
Michal Harel (talk | contribs) No edit summary |
Michal Harel (talk | contribs) No edit summary |
||
| (13 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
<StructureSection load=' | <StructureSection load='2exs' size='350' side='right' caption='Structure of tryptophan RNA-binding attenuation protein complex with Trp (PDB code [[2exs]]).' scene=''> | ||
== Function == | == Function == | ||
'''Tryptophan RNA-binding attenuation protein''' (TRAP) is a tryptophan-activated RNA-binding protein which regulates the expression of the tryptophan biosynthesis genes | '''Tryptophan RNA-binding attenuation protein''' (TRAP) or '''transcription attenuation protein MtrB''' is a tryptophan-activated RNA-binding protein which regulates the expression of the tryptophan biosynthesis genes. Down regulation of Trp biosynthesis is observed upon binding of 11 tryptophan molecules to TRAP<ref>PMID:14687568</ref>. | ||
== Structural highlights == | == Structural highlights == | ||
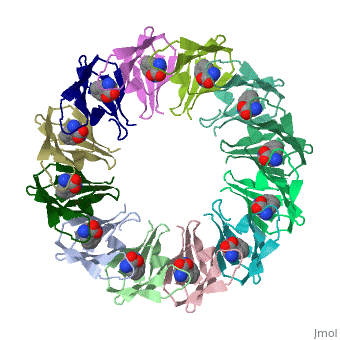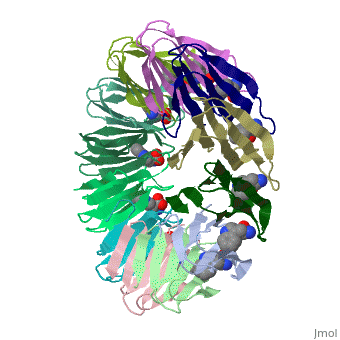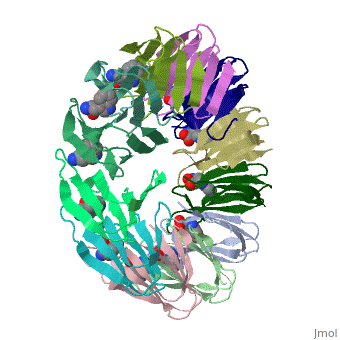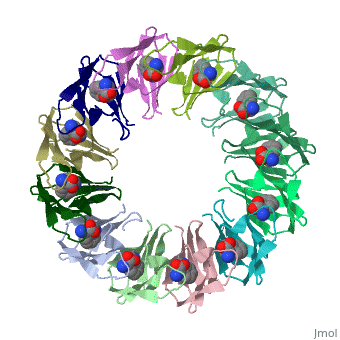
TRAP is composed of 11 identical subunits arranged in a <scene name='59/595823/Cv/4'>symmetrical ring</scene>. The <scene name='59/595823/Cv/3'>active site of TRAP</scene><ref>PMID:16698553</ref>. Water molecules are shown as red spheres. | |||
</StructureSection> | |||
== 3D Structures of tryptophan RNA-binding attenuation protein == | == 3D Structures of tryptophan RNA-binding attenuation protein == | ||
Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
{{#tree:id=OrganizedByTopic|openlevels=0| | |||
*Tryptophan RNA-binding attenuation protein | |||
**[[3aqd]] – GsTRAP – ''Geobacillus stearothermophilus''<br /> | |||
*Tryptophan RNA-binding attenuation protein binary complex with Trp | |||
**[[2zdo]], [[2exs]], [[2ext]], [[2zcz]], [[2zd0]], [[3zzs]], [[1qaw]] – GsTRAP + Trp<br /> | |||
**[[5eew]], [[5eex]], [[5eey]], [[5eez]], [[5ef0]], [[5ef1]], [[5ef2]], [[5ef3]] – GsTRAP + Trp – radiation damage<br /> | |||
**[[1wap]], [[3zzq]] – BsTRAP + Trp – ''Bacillus subtilis''<br /> | |||
**[[4b27]] – BsTRAP (mutant) + Trp <br /> | |||
**[[3zte]] – TRAP + Trp – ''Bacillus licheniformis''<br /> | |||
**[[3zzl]] – TRAP + Trp – ''Bacillus halodurans''<br /> | |||
*Tryptophan RNA-binding attenuation protein ternary complex with Trp | |||
**[[1c9s]], [[1gtf]], [[1gtn]], [[1utd]], [[4v4f]] – GsTRAP + RNA + Trp <br /> | |||
**[[2zp8]], [[2zp9]] – GsTRAP + TRAP inhibitory protein + Trp <br /> | |||
}} | |||
== References == | == References == | ||
<references/> | <references/> | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Latest revision as of 13:31, 21 December 2020
FunctionTryptophan RNA-binding attenuation protein (TRAP) or transcription attenuation protein MtrB is a tryptophan-activated RNA-binding protein which regulates the expression of the tryptophan biosynthesis genes. Down regulation of Trp biosynthesis is observed upon binding of 11 tryptophan molecules to TRAP[1]. Structural highlightsTRAP is composed of 11 identical subunits arranged in a . The [2]. Water molecules are shown as red spheres. |
| ||||||||||
3D Structures of tryptophan RNA-binding attenuation protein3D Structures of tryptophan RNA-binding attenuation protein
Updated on 21-December-2020
ReferencesReferences
- ↑ Li PT, Gollnick P. Characterization of a trp RNA-binding attenuation protein (TRAP) mutant with tryptophan independent RNA binding activity. J Mol Biol. 2004 Jan 16;335(3):707-22. PMID:14687568 doi:http://dx.doi.org/10.1016/S0022283603013846
- ↑ Heddle JG, Yokoyama T, Yamashita I, Park SY, Tame JR. Rounding up: Engineering 12-membered rings from the cyclic 11-mer TRAP. Structure. 2006 May;14(5):925-33. PMID:16698553 doi:http://dx.doi.org/10.1016/j.str.2006.03.013