Help:Making animations for Powerpoint: Difference between revisions
Eric Martz (talk | contribs) |
Eric Martz (talk | contribs) No edit summary |
||
| (21 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
Any molecular scene in Proteopedia (or any Jmol website) can be converted to a rocking or spinning animation for presentations, such as Microsoft Powerpoint, free Google Slides, or free Libre Office. | Any molecular scene in Proteopedia (or any Jmol website) can be converted to a rocking or spinning animation for presentations, such as Microsoft Powerpoint, free Google Slides, or free Libre Office. | ||
Proteopedia has a built-in mechanism to make a presentation-ready animation. Simply click on ''Export Animated Image'' below the molecular scene. You can also export a static image there. | |||
In February, 2019, Proteopedia's mechanism is somewhat limited. The animations are jerky and will perhaps be smaller than you wish. An alternative is to make an animation of a Proteopedia scene using [[FirstGlance in Jmol]]. | |||
Making presentation-ready animations in ''FirstGlance in Jmol'' is quick and easy, and the animations will rotate more smoothly and can be larger (hence clearer/sharper). | |||
If you want to animate a molecular scene that you first obtain in FirstGlance in Jmol, then you don't need the methods below. The capability is built into FirstGlance: see the snapshot in step #7 below (in Method I). The methods below are only for generating an animation in FirstGlance from a scene in Proteopedia. | |||
==Demonstration Examples== | ==Demonstration Examples== | ||
| Line 10: | Line 16: | ||
This method will make a presentation-ready rocking or spinning animation of a fixed molecular scene. See examples linked above. It will not convert complex Jmol animations such as [[Morphs]] or those in [[Molecular Playground/HIV Protease Inhibitor]] and [[Molecular Playground/Tamiflu]] into presentation-ready animations. However, morphs can be made presentation-ready using [http://polyview.cchmc.org/polyview3d.html Polyview-3D]. An example is the last slide in this downloadable [http://bioinformatics.org/firstglance/fgij/ppt/polyview-3d-examples.ppt Powerpoint demonstration of Polyview-3D animations]. If you need help with a morph, please email [[Image:Contact-email.png]]. | This method will make a presentation-ready rocking or spinning animation of a fixed molecular scene. See examples linked above. It will not convert complex Jmol animations such as [[Morphs]] or those in [[Molecular Playground/HIV Protease Inhibitor]] and [[Molecular Playground/Tamiflu]] into presentation-ready animations. However, morphs can be made presentation-ready using [http://polyview.cchmc.org/polyview3d.html Polyview-3D]. An example is the last slide in this downloadable [http://bioinformatics.org/firstglance/fgij/ppt/polyview-3d-examples.ppt Powerpoint demonstration of Polyview-3D animations]. If you need help with a morph, please email [[Image:Contact-email.png]]. | ||
==Procedure== | |||
= | |||
Display the desired molecular scene. Then: | Display the desired molecular scene. Then: | ||
<ol> | <ol> | ||
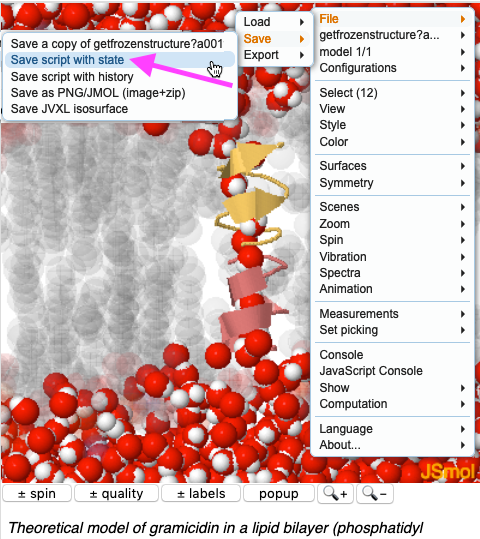
<li>Right click near the top of JSmol to open the JSmol menu. | <li>Right click (Macs: Ctrl-click) near the top of JSmol to open the JSmol menu. Touch <i>File</i>, then <i>Save</i>, then click on '''Save script with state'''.</li> | ||
[[Image:Jsmol-menu- | [[Image:Jsmol-menu-save-script-with-state.png|center]] | ||
<li>Change the file name if you wish. The conventional file type is '''.spt''' but the file name does not matter. Complete the file saving dialogs. The file will be saved in the location specified in the preferences of the browser you are using.</li> | |||
<li> | |||
<li> | <li>Right click [http://firstglance.jmol.org FirstGlance in Jmol] to open it in a new tab/window, so that this procedure remains available. | ||
<li>Display '''4QXX''' in FirstGlance. (This simple peptide avoids some problems.) | <li>Display '''4QXX''' in FirstGlance. (This simple peptide avoids some problems.) | ||
</li> | </li> | ||
<li>Drag the | <li>Drag the "script with state" file that you saved above and drop it into JSmol (drop on top of the molecule) in FirstGlance. The molecular scene from Proteopedia should appear in less than one minute. | ||
<ul> | <ul> | ||
<li> | <li> | ||
You may use the Spin, Background, Quality and Zoom buttons. Clicking other things in FirstGlance may ruin your scene, but then you can easily restore it by dropping the | You may use the Spin, Background, Quality and Zoom buttons. Clicking other things in FirstGlance may ruin your scene, but then you can easily restore it by dropping the script file in again. | ||
</li> | </li> | ||
</ul> | </ul> | ||
| Line 52: | Line 41: | ||
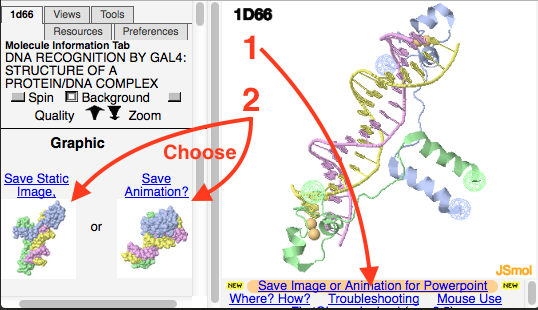
<li>Click on ''Save Image or Animation for Powerpoint'', and choose ''Save Animation''.</li> | <li>Click on ''Save Image or Animation for Powerpoint'', and choose ''Save Animation''.</li> | ||
[[Image:Fgij-arrows-12.png | [[Image:Fgij-arrows-12.png|center]] | ||
<li>Follow the instructions that appear | <li>Follow the instructions that appear to save your animation GIF file. | ||
</li> | </li> | ||
| Line 132: | Line 50: | ||
</ol> | </ol> | ||
==Animation Kit== | |||
FirstGlance in Jmol's built-in animation maker also has limitations on the size of the animation, particularly if you want 360° spinning. To enable you to make such animations as large and crisp as you like, FirstGlance provides a [https://bioinformatics.org/firstglance/fgij/animation_kit_instructions.pdf downloadable Animation Kit] with complete [https://bioinformatics.org/firstglance/fgij/animation_kit_instructions.pdf instructions]. | |||
==Feedback== | ==Feedback== | ||
Problems? Unclear? Please send feedback or questions to [[Image:Contact-email.png]]. | Problems? Unclear? Please send feedback or questions to [[Image:Contact-email.png]]. | ||
Latest revision as of 21:23, 9 August 2020
Any molecular scene in Proteopedia (or any Jmol website) can be converted to a rocking or spinning animation for presentations, such as Microsoft Powerpoint, free Google Slides, or free Libre Office.
Proteopedia has a built-in mechanism to make a presentation-ready animation. Simply click on Export Animated Image below the molecular scene. You can also export a static image there.
In February, 2019, Proteopedia's mechanism is somewhat limited. The animations are jerky and will perhaps be smaller than you wish. An alternative is to make an animation of a Proteopedia scene using FirstGlance in Jmol.
Making presentation-ready animations in FirstGlance in Jmol is quick and easy, and the animations will rotate more smoothly and can be larger (hence clearer/sharper).
If you want to animate a molecular scene that you first obtain in FirstGlance in Jmol, then you don't need the methods below. The capability is built into FirstGlance: see the snapshot in step #7 below (in Method I). The methods below are only for generating an animation in FirstGlance from a scene in Proteopedia.
Demonstration ExamplesDemonstration Examples
LimitationsLimitations
This method will make a presentation-ready rocking or spinning animation of a fixed molecular scene. See examples linked above. It will not convert complex Jmol animations such as Morphs or those in Molecular Playground/HIV Protease Inhibitor and Molecular Playground/Tamiflu into presentation-ready animations. However, morphs can be made presentation-ready using Polyview-3D. An example is the last slide in this downloadable Powerpoint demonstration of Polyview-3D animations. If you need help with a morph, please email ![]() .
.
ProcedureProcedure
Display the desired molecular scene. Then:
- Right click (Macs: Ctrl-click) near the top of JSmol to open the JSmol menu. Touch File, then Save, then click on Save script with state.
- Change the file name if you wish. The conventional file type is .spt but the file name does not matter. Complete the file saving dialogs. The file will be saved in the location specified in the preferences of the browser you are using.
- Right click FirstGlance in Jmol to open it in a new tab/window, so that this procedure remains available.
- Display 4QXX in FirstGlance. (This simple peptide avoids some problems.)
- Drag the "script with state" file that you saved above and drop it into JSmol (drop on top of the molecule) in FirstGlance. The molecular scene from Proteopedia should appear in less than one minute.
- You may use the Spin, Background, Quality and Zoom buttons. Clicking other things in FirstGlance may ruin your scene, but then you can easily restore it by dropping the script file in again.
- Click on Save Image or Animation for Powerpoint, and choose Save Animation.
- Follow the instructions that appear to save your animation GIF file.
- Drag the saved GIF file and drop it into a slide in your presentation.
Animation KitAnimation Kit
FirstGlance in Jmol's built-in animation maker also has limitations on the size of the animation, particularly if you want 360° spinning. To enable you to make such animations as large and crisp as you like, FirstGlance provides a downloadable Animation Kit with complete instructions.