ModG: Difference between revisions
Michal Harel (talk | contribs) No edit summary |
Michal Harel (talk | contribs) No edit summary |
||
| Line 1: | Line 1: | ||
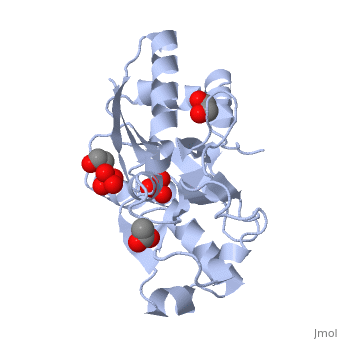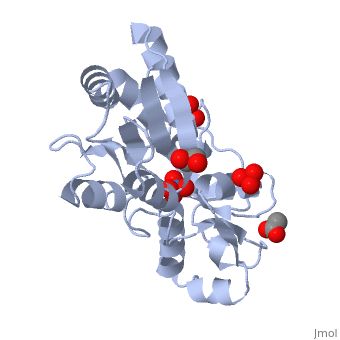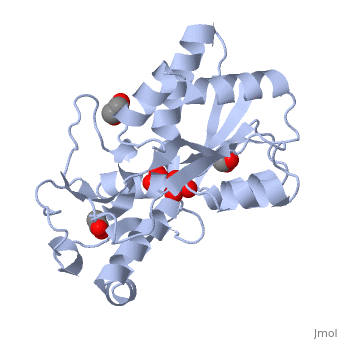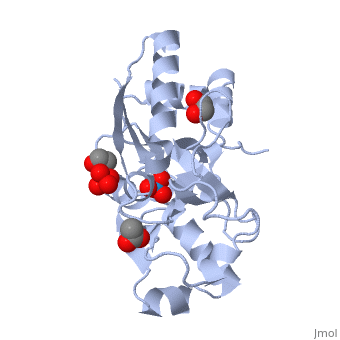
<StructureSection load='1atg' size='350' side='right' caption='Molybdate-binding protein complex with tungstate, ethylene glycol, acetate and sulfate (PDB entry [[1atg]])' scene=''> | <StructureSection load='1atg' size='350' side='right' caption='Molybdate-binding protein complex with tungstate, ethylene glycol, acetate and sulfate (PDB entry [[1atg]])' scene=''> | ||
Latest revision as of 09:49, 24 July 2017
Introduction to ModGModG is a cytoplasmic molybdate-binding protein exclusive to the aerobic nitrogen-fixer Azobacter vinelandii.[1] Molybdate is a molybdenum oxyanion (MoO42-). The group 6 element molybdenum is required by many enzymes that catalyze reactions associated with carbon, nitrogen, or sulfur metabolism.[2] It is also part of the cofactor of the molybdoenzyme ModG. Not surprisingly, studies have linked ModG to molybdenum homeostasis within the cell.[1] StructureThe molybdoenzyme is a homotrimer.[1] It can bind up to 8 molybdate molecules between 4 different types of on subunit interfaces (BS1, BS1’, BS2, and BS2’). Binding site 1 and binding site 2 are found at opposite ends of the protein; binding site 1’ and binding site 2’ are found off-axis near like ends.[3] The sites are connected by hydrogen bonds and thus a cooperative binding mechanism has been proposed for ModG whereby ligand engagement with type 2 sites induces conformational changes to asparagine residues at type 1 sites, reading the site for ligand interactions.[4] The structure of ModG was solved by Delarbe et al. using multi-wavelength anomalous dispersion (MAD).[5] Crystallization required salt-free conditions established with polyethylene glycol (PEG), at which point the authors solved the PEG crystal form using molecular replacement.[5] The protein is made up of 3 identical subunits that have 67 amino acid pairs and are 14.3 kDa in size.[1] In trimer form the subunits are perpendicular to each other and intersect at the 3-fold axis (as seen in the crystallized form).[3] Each subunit is composed of two β-barrel domains (Domain I and Domain II) that each feature a short 310-helix.[3] Each β-barrel domain includes five antiparallel β-strands arranged in a motif that is capped by two-turn α-helices.[3] The folding arrangement of the ModG domains is that of an oligomer-binding (OB) fold, characteristic of toxins and other intracellular oxyanion-binding proteins.[3] In trimerization, N and C termini (both found in Domain I) of a subunit interact with the β5-β6 loop (Domain II) of the adjoining subunit by sharing antiparallel β-sheets. Approximately 40% of monomer surface area is buried when the protein is in trimer form.[1]
Properties and binding affinityAbout 60% of each subunit interface is (40% is polar); furthermore, there is an unequal charge distribution that is attributed to the presence of 4 side chains.[2][4] These include: Lys60, Lys132, Arg6, and Arg78.[1] The are both found on the type 2 binding sites and are entirely buried in the protein; the are partly exposed.[1] This leaves the interface particularly electropositive; electrostatic repulsion is balanced by several favorable interactions that include the formation of 24 hydrogen bonds, 2 salt bridges and approximately 24% of total hydrophobic surface buried (per subunit).[4] The trimer is further stabilized by the binding of an oxyanion which would have a neutralizing effect.[3] Type 1 binding sites utilize hydrogen bonds and interactions with uncharged polar residues to bind molybdate.[4] The tight pocket volume and rigidity of the site suggest that there is high selectivity for molybdate (or tungstate because ModG cannot differentiate between the two oxyanions).[4] Type 2 sites are comparatively larger in pocket volume, supple, and feature electropositive lysine residues that would contribute to high affinity for negatively charged oxyanions like molybdate.[4] Because the type 2 sites are larger and more flexible, there would be less specificity for molybdate and thus other oxyanions such as phosphate would likely compete for binding.[3][4]
Function in BacteriaThe ability of A. vinelandii to differentiate between molybdate and other oxyanions is of particular interest to researchers.[1][2][5] Phosphate is distinguished from molybdate by the divergence in protonation states: phosphate is protonated at physiological pH whereas molybdate is not.[2] The distinction between sulfate and molybdate is reliant upon differences in ligand size.[2] Interestingly, molybdate permeases are not able to distinguish between tungstate and molybdate.[1][2] Regulation of these intracellular metabolites is attributed to other molybdate-binding proteins collectively known as molbindins.[1][5]
FateModG is eventually degraded and incorporated into molybdopterin, a cofactor of molybdenum enzymes, or the iron-molybdenum cofactor of a nitrogenase enzyme.[1][2] Currently there is little known about cofactor biosynthesis involving the ModG protein.[1] |
| ||||||||||
3D structures of ModG3D structures of ModG
1h9j – AvModG + phosphate + molybdate – Azotobacter vinelandii
1atg – AvModG + tungstate + sulfate + acetate
1h9k - AvModG + tungstate + phosphate
1h9m - AvModG + molybdate
ReferencesReferences
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 Delarbre L, Stevenson CE, White DJ, Mitchenall LA, Pau RN, Lawson DM. Two crystal structures of the cytoplasmic molybdate-binding protein ModG suggest a novel cooperative binding mechanism and provide insights into ligand-binding specificity. J Mol Biol. 2001 May 18;308(5):1063-79. PMID:11352591 doi:10.1006/jmbi.2001.4636
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 Yang ZY, Dean DR, Seefeldt LC. Molybdenum nitrogenase catalyzes the reduction and coupling of CO to form hydrocarbons. J Biol Chem. 2011 Mar 28. PMID:21454640 doi:10.1074/jbc.M111.229344
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 Lawson DM, Williams CE, Mitchenall LA, Pau RN. Ligand size is a major determinant of specificity in periplasmic oxyanion-binding proteins: the 1.2 A resolution crystal structure of Azotobacter vinelandii ModA. Structure. 1998 Dec 15;6(12):1529-39. PMID:9862806
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 4.6 Mouncey NJ, Mitchenall LA, Pau RN. Mutational analysis of genes of the mod locus involved in molybdenum transport, homeostasis, and processing in Azotobacter vinelandii. J Bacteriol. 1995 Sep;177(18):5294-302. PMID:7665518
- ↑ 5.0 5.1 5.2 5.3 Williams CE, White DJ, Delarbre L, Mitchenall LA, Pau RN, Lawson DM. Crystallization and preliminary X-ray studies on the molbindin ModG from Azotobacter vinelandii. Acta Crystallogr D Biol Crystallogr. 1999 Jul;55(Pt 7):1356-8. PMID:10393312
See alsoSee also
This page originally authored by Corbin Black