Sandbox 3: Difference between revisions
Eran Hodis (talk | contribs) No edit summary |
David Canner (talk | contribs) |
||
| (16 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
[[Image:1ema.gif|thumb|left|350px|Green fluorescent protein (1ema)]] | |||
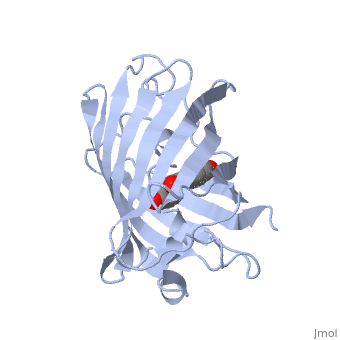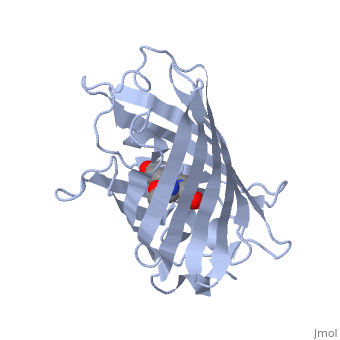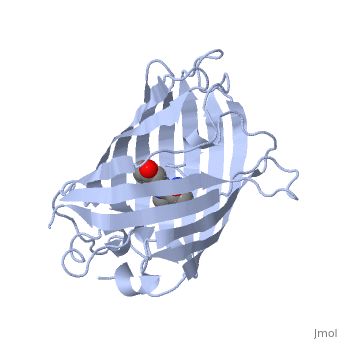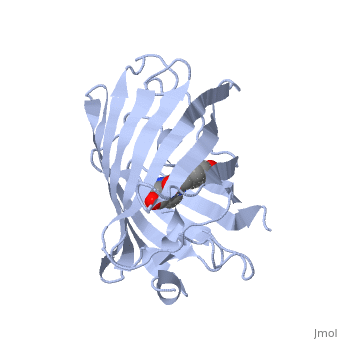
Green fluorescent protein ('''GFP'''), originally isolated from the jellyfish Aequorea victoria (PDB entry [[1ema]]), fluorsceses green (509nm) when exposed to blue light (395nm and 475nm). It is one of the most important proteins used in biological research because it can be used to tag otherwise invisible gene products of interest and thus observe their existence, location and movement. | |||
== Exploring the Structure == | == Exploring the Structure == | ||
<applet load='1ema' size='300' frame='true' align='right' caption='Insert caption here' /> | |||
GFP is a beta barrel protein with 11 beta sheets. It is a 26.9kDa protein made up of 238 amino acids. The <scene name='Sandbox_3/1ema_chromophore/1'>chromophore</scene>, responsible for the fluorescent properties of the protein, is buried inside the beta barrel as part of the central alpha helix passing through the barrel. The chromophore forms via spontaneous cyclization and oxidation of three residues in the central alpha helix: -Thr65 (or Ser65)-Tyr66-Gly67. This cyclization and oxidation creates the chromophore's five-membered ring via a new bond between the threonine and the glycine residues.<ref>PMID:8703075</ref> | |||
<references/> | |||
<br /> | |||
Follow this link: [http://www.proteopedia.org/wiki/workshop/GFP_sample.html Click Here] | |||
Latest revision as of 17:45, 19 May 2012

Green fluorescent protein (GFP), originally isolated from the jellyfish Aequorea victoria (PDB entry 1ema), fluorsceses green (509nm) when exposed to blue light (395nm and 475nm). It is one of the most important proteins used in biological research because it can be used to tag otherwise invisible gene products of interest and thus observe their existence, location and movement.
Exploring the StructureExploring the Structure
|
GFP is a beta barrel protein with 11 beta sheets. It is a 26.9kDa protein made up of 238 amino acids. The , responsible for the fluorescent properties of the protein, is buried inside the beta barrel as part of the central alpha helix passing through the barrel. The chromophore forms via spontaneous cyclization and oxidation of three residues in the central alpha helix: -Thr65 (or Ser65)-Tyr66-Gly67. This cyclization and oxidation creates the chromophore's five-membered ring via a new bond between the threonine and the glycine residues.[1]
Follow this link: Click Here