Sandbox Reserved 470: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
No edit summary |
||
| (13 intermediate revisions by the same user not shown) | |||
| Line 4: | Line 4: | ||
==Introduction== | ==Introduction== | ||
---- | ---- | ||
'''[[Glyceraldehyde-3-phosphate Dehydrogenase]]''' | *'''[[Glyceraldehyde-3-phosphate Dehydrogenase]]''' ([http://www.rcsb.org/pdb/home/home.do RSCB] (PDB)used:3dpg) | ||
<Structure load='3gpd' size='350' frame='true' align='right' caption='Glyceraldehyde-3-phosphate Dehydrogenase' scene='Insert optional scene name here' />(abbreviated as GAPDH or the less common G3PDH) (EC 1.2.1.12) ~37kDa is a well-known [http://en.wikipedia.org/wiki/Enzyme enzyme] which catalyzes the sixth step of [[glycolysis]], a reversible cytosolic process in [http://en.wikipedia.org/wiki/Eukaryote eukaryotes] which involves the breakdown of glucose for energy and carbon molecules. Along with its role in glycolysis and [http://en.wikipedia.org/wiki/Gluconeogenesis gluconeogenesis], recent research has determined that GAPDH is actually a multifunctional protein, as it has numerous defined, non-metabolic functions involved in multiple subcellular processes including [http://en.wikipedia.org/wiki/Transcription transcription] activation, ER to Golgi transportation, transcriptional control of histone [http://en.wikipedia.org/wiki/Gene_Expression gene expression], nuclear membrane fusion, neuronal initiation of [http://en.wikipedia.org/wiki/Apoptosis apoptosis], recognizing fraudulently incorporated nucleotides in DNA, and maintaining [http://en.wikipedia.org/wiki/Telomere telomere] structures. Research also shows that it possibly has a direct involvement in cellular phenotype of human [http://en.wikipedia.org/wiki/Neurodegeneration neurodegenerative] disorders, especially those characterized by expansion of [http://en.wikipedia.org/wiki/CAG_Triplet_Repeat_Disorders CAG repeats]. | <Structure load='3gpd' size='350' frame='true' align='right' caption='Glyceraldehyde-3-phosphate Dehydrogenase' scene='Insert optional scene name here' />(abbreviated as GAPDH or the less common G3PDH) (EC 1.2.1.12) ~37kDa is a well-known [http://en.wikipedia.org/wiki/Enzyme enzyme] which catalyzes the sixth step of [[glycolysis]], a reversible cytosolic process in [http://en.wikipedia.org/wiki/Eukaryote eukaryotes] which involves the breakdown of glucose for energy and carbon molecules. Along with its role in glycolysis and [http://en.wikipedia.org/wiki/Gluconeogenesis gluconeogenesis], recent research has determined that GAPDH is actually a multifunctional protein, as it has numerous defined, non-metabolic functions involved in multiple subcellular processes including [http://en.wikipedia.org/wiki/Transcription transcription] activation, ER to Golgi transportation, transcriptional control of histone [http://en.wikipedia.org/wiki/Gene_Expression gene expression], nuclear membrane fusion, neuronal initiation of [http://en.wikipedia.org/wiki/Apoptosis apoptosis], microtubule bundling, phosphotransferase activity, prostate cancer progression, recognizing fraudulently incorporated nucleotides in DNA, and maintaining [http://en.wikipedia.org/wiki/Telomere telomere] structures. Research also shows that it possibly has a direct involvement in cellular phenotype of human [http://en.wikipedia.org/wiki/Neurodegeneration neurodegenerative] disorders, especially those characterized by expansion of [http://en.wikipedia.org/wiki/CAG_Triplet_Repeat_Disorders CAG repeats]. | ||
==Structure== | ==Structure== | ||
---- | ---- | ||
GAPDH has two domains, a GAPDH-like, C-terminal domain, and an NAD Binding Domain. | *While GAPDH possesses four cysteine residues, its <scene name='Sandbox_Reserved_470/Active_site_gapdh/1'>active site</scene> is located around <scene name='Sandbox_Reserved_470/Cysteine_active_site/1'>Cysteine 149</scene>, which is responsible for inhibition of glycolysis and fermentation by iodoacetic acid. '''<ref>''' Boyer, Paul D.. "The Enzymes - Paul D. Boyer - Google Books." Google Books. N.p., n.d. Web. 1 May 2012. '''[<http://books.google.com/books?id=r6SRFsxYFYwC&pg=PA20&lpg=PA20&dq=GAPDH+cysteine+149&source=bl&ots=0CWwdduzGB&sig=Lb8PtWq7EGdwyvdAnFI_rHYG3tU&hl=en&sa=X&ei=2PyhT-WtHJSy8QTjtsnTCA&ved=0CDMQ6AEwAg#v=onepage&q=GAPDH%20cysteine%20149&f=false>]'''. '''</ref>''' | ||
*GAPDH has two domains, a GAPDH-like, C-terminal domain, and an NAD Binding Domain. | |||
:The <scene name='Sandbox_Reserved_470/Nad_binding_domain/3'>NAD (Nucleotide) Binding Domain </scene> is an Alpha Beta 3-Layer(α-β-α) Sandwich, including amino acids 1-138 and 301-340 on chains O and Q . Its [http://en.wikipedia.org/wiki/CATH CATH] reports a Rossmann fold and a classification as an [http://en.wikipedia.org/wiki/Oxidoreductase oxidoreductase]. | |||
:The <scene name='Sandbox_Reserved_470/Gapdh-like_c-terminal_domain/3'>GAPDH-like, C-terminal domain</scene>GAPDH-like, C-terminal domain (Catalytic domain) is an Alpha Beta 2-Layer(α-β) Sandwich, including amino acids 139 - 300 on chains O and Q . Its CATH reports a holo-D-Glyceraldehyde-3-Phosphate Dehydrogenase, (domain 2) and a classification as an oxidoreductase (aldehyde(D)-NAD(A)). | |||
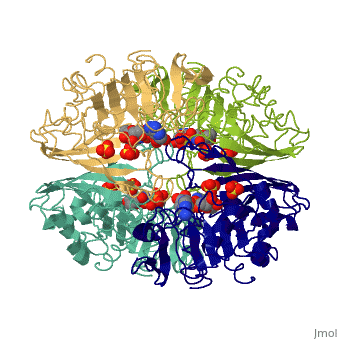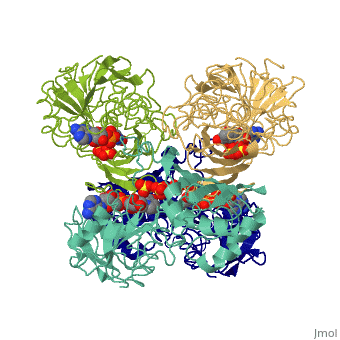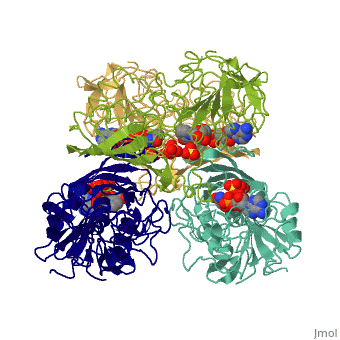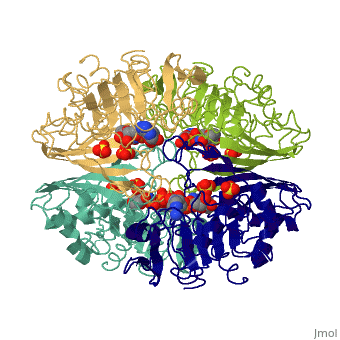
*Depending on the environment, including variations in temperature and acidity, GAPDH can have different structural qualities and physical properties. In some cases, it is observed as a twinned structure with an α/β barrel. '''<ref>'''Watson, H.C., and J.C. Campbell. "Twinning In Crystals Of Human Skeletal Muscle D-Glyceraldehyde-3-Phosphate Dehydrogenase." RCSB Protein Data Bank. N.p., 27 Oct. 1983. Web. 28 Apr. 2012. '''[<www.rcsb.org/pdb/explore/explore.do?structureId=3gpd>]'''. '''</ref>''' | |||
[[Image:GAPDH homotetramer 2.png|300 px]] | |||
==Importance== | ==Importance== | ||
---- | ---- | ||
'''Role in Glycolysis:''' | ==='''Role in Glycolysis:'''=== | ||
====The Steps:==== | |||
:In two coupled steps, | :In two coupled steps, | ||
[[Image:GAPDH_PGK-rxn.gif|400 px]] | [[Image:GAPDH_PGK-rxn.gif|400 px]] | ||
GAPDH catalyzes the conversion of [http://en.wikipedia.org/wiki/Glyceraldehyde-3-phosphate glyceraldyhyde-3-phosphate] at carbon 1 to [http://en.wikipedia.org/wiki/1,3-bisphosphoglycerate 1,3-bisphosphoglycerate] (1,3-BPG). | GAPDH catalyzes the conversion of [http://en.wikipedia.org/wiki/Glyceraldehyde-3-phosphate glyceraldyhyde-3-phosphate] at carbon 1 to [http://en.wikipedia.org/wiki/1,3-bisphosphoglycerate 1,3-bisphosphoglycerate] (1,3-BPG). | ||
====The Reactions:==== | |||
:The reaction involving the GAPDH enzyme combines phosphorylation with oxidation in an overall [http://en.wikipedia.org/wiki/Endergonic endergonic] reaction. (ΔG°'=+6.3 kJ/mol (+1.5 kcal/mol)) First, the oxidation of glyceraldyhyde-3-phosphate to D-glycerate 1,3-bisphosphate takes place, in which an aldehyde is converted to carboxylic acid ((ΔG°'=-50 kJ/mol (-12 kcal/mol))and NAD+ ([http://en.wikipedia.org/wiki/Nicotinamide_Adenine_Dinucleotide Nicotinamide adenine dinucleotide]), an important co-factor and [http://en.wikipedia.org/wiki/Ligand ligand] found bound to the <scene name='Sandbox_Reserved_470/Active_site_gapdh/1'>active site</scene> of GAPDH, is simultaneously reduced endergonically to NADH. This oxidation reaction is required for the initiation of the second reaction because it is highly [http://en.wikipedia.org/wiki/Exergonic exergonic] and thus drives the endergonic second reaction ((ΔG°'=+50 kJ/mol (+12 kcal/mol)). In the second reaction a molecule of inorganic phosphate is transferred to a GAP intermediate to form a product with a high potential to transfer phosphates, 1,3-bisphosphoglycerate. | :The reaction involving the GAPDH enzyme combines phosphorylation with oxidation in an overall [http://en.wikipedia.org/wiki/Endergonic endergonic] reaction. (ΔG°'=+6.3 kJ/mol (+1.5 kcal/mol)) First, the oxidation of glyceraldyhyde-3-phosphate to D-glycerate 1,3-bisphosphate takes place, in which an aldehyde is converted to carboxylic acid ((ΔG°'=-50 kJ/mol (-12 kcal/mol))and NAD+ ([http://en.wikipedia.org/wiki/Nicotinamide_Adenine_Dinucleotide Nicotinamide adenine dinucleotide]), an important co-factor and [http://en.wikipedia.org/wiki/Ligand ligand] found bound to the <scene name='Sandbox_Reserved_470/Active_site_gapdh/1'>active site</scene> of GAPDH, is simultaneously reduced endergonically to NADH. This oxidation reaction is required for the initiation of the second reaction because it is highly [http://en.wikipedia.org/wiki/Exergonic exergonic] and thus drives the endergonic second reaction ((ΔG°'=+50 kJ/mol (+12 kcal/mol)). In the second reaction a molecule of inorganic phosphate is transferred to a GAP intermediate to form a product with a high potential to transfer phosphates, 1,3-bisphosphoglycerate. | ||
====The Mechanism:==== | |||
:The mechanism of GAPDH is mainly dependent the [http://en.wikipedia.org/wiki/Thiol thiol] functional group on a cysteine residue which lies within the <scene name='Sandbox_Reserved_470/Active_site_gapdh/1'>active site</scene>. This important thiol group acts as a [http://en.wikipedia.org/wiki/Ncleophile nucleophile], attacking the carbon on the aldehyde functional group of the [http://en.wikipedia.org/wiki/Substrate substrate], glyceraldehyde 3-phosphate, after it binds to GAPDH. The creation of a [http://en.wikipedia.org/wiki/Thiohemiacetal thiohemiacetal] intermediate occurs from this oxidative reaction, which then loses a hydride to NAD+, which is also bound nearby, forming NADH and a carboxyl group from the [http://en.wikipedia.org/wiki/Aldehyde aldehyde]. Conservation of energy is maintained through this thioester linkage to the active site cysteine. The GAP intermediate is then used in the second step, where it is important to notice that without GAPDH's use of covalent catalysis, the energy barrier of the reaction would be too high and the reaction would be too slow for living organisms. | :The mechanism of GAPDH is mainly dependent the [http://en.wikipedia.org/wiki/Thiol thiol] functional group on a cysteine residue, of the 3-phosphoglycerate amino acid biosynthetic family, which lies within the <scene name='Sandbox_Reserved_470/Active_site_gapdh/1'>active site</scene>. This important thiol group acts as a [http://en.wikipedia.org/wiki/Ncleophile nucleophile], attacking the carbon on the aldehyde functional group of the [http://en.wikipedia.org/wiki/Substrate substrate], glyceraldehyde 3-phosphate, after it binds to GAPDH. The creation of a [http://en.wikipedia.org/wiki/Thiohemiacetal thiohemiacetal] intermediate occurs from this oxidative reaction, which then loses a hydride to NAD+, which is also bound nearby, forming NADH and a carboxyl group from the [http://en.wikipedia.org/wiki/Aldehyde aldehyde]. Conservation of energy is maintained through this thioester linkage to the active site cysteine. The GAP intermediate is then used in the second step, where it is important to notice that without GAPDH's use of covalent catalysis, the energy barrier of the reaction would be too high and the reaction would be too slow for living organisms. | ||
---- | ---- | ||
'''Other roles:''' | ==='''Other roles:'''=== | ||
*The importance of GAPDH in transcription was discovered by Zheng ''et. al''. in 2003. It was found that OCA's transcriptional coactivator complex contains GAPDH, which moves between the cytosol and nucleus and may link the metabolic state to gene transcription. | *The importance of GAPDH in transcription was discovered by Zheng ''et. al''. in 2003. It was found that OCA's transcriptional coactivator complex contains GAPDH, which moves between the cytosol and nucleus and may link the metabolic state to gene transcription. | ||
*In 2005 the initiation of apoptosis was shown to be mediated by GAPDH by Hara ''et. al.'' when it was found to bind to DNA like it does in transcription activation. | *In 2005 the initiation of apoptosis was shown to be mediated by GAPDH by Hara ''et. al.'' when it was found to bind to DNA like it does in transcription activation. | ||
| Line 32: | Line 35: | ||
*It was also found that GAPDH is plays a role in certain neruodegenerative disorders, as is able to find stretches which are encoded by the gene's CAG repeats and bind to the gene products formed from disorders such as [http://en.wikipedia.org/wiki/Huntington's_disease Huntington's disease], [http://en.wikipedia.org/wiki/Alzheimer's_disease Alzheimer’s disease], [http://en.wikipedia.org/wiki/Parkinson's_disease Parkinson’s disease] and [http://en.wikipedia.org/wiki/Machado-Joseph_disease Machado-Joseph disease]. | *It was also found that GAPDH is plays a role in certain neruodegenerative disorders, as is able to find stretches which are encoded by the gene's CAG repeats and bind to the gene products formed from disorders such as [http://en.wikipedia.org/wiki/Huntington's_disease Huntington's disease], [http://en.wikipedia.org/wiki/Alzheimer's_disease Alzheimer’s disease], [http://en.wikipedia.org/wiki/Parkinson's_disease Parkinson’s disease] and [http://en.wikipedia.org/wiki/Machado-Joseph_disease Machado-Joseph disease]. | ||
---- | ---- | ||
'''Use in the lab:''' | ==='''Use in the lab:'''=== | ||
*Overall, GAPDH is | ====Quantitation:==== | ||
*Overall, GAPDH is one of the most common “housekeeping genes” and is found in high levels in tissues and cells so it is commonly used in biological research as a loading control in quantification techniques such as [http://en.wikipedia.org/wiki/Lowry_assay Lowry Assays], [http://en.wikipedia.org/wiki/Bradford_assay Bradford Assays], [http://en.wikipedia.org/wiki/Isoelectric_focusing Isoelectric Focusing], 2D-PAGE, [http://en.wikipedia.org/wiki/Size-exclusion_chromatography Size Exclusion Chromatography] (SEC), as well as [http://en.wikipedia.org/wiki/SDS-PAGE 1-D and 2-D SDS-PAGE] when it is conjugated to a known amount of biotin or BODIPY fluorophore to create a standard. A good method of quantitation lies under proteomics, which concompasses a variation of techniques including Differential in-gel Electrophoresis (DIGE), [http://en.wikipedia.org/wiki/SILAC SILAC] (for Metabolic Labeling), [http://en.wikipedia.org/wiki/ITRAQ iTRAQ] (for Chemical Labeling). | |||
====Qualification and Analysis:==== | |||
*Analysis of GAPDH can give much useful information when comparing proteins and substances in an experiment, thus some techniques used include [http://en.wikipedia.org/wiki/Affinity_chromatography Affinity Chromatography], [http://en.wikipedia.org/wiki/Ion_Exchange_chromatography Ion Exchange Chromatography], [http://en.wikipedia.org/wiki/Reversed-phase_liquid_chromatography Reversed Phase LC]. Often times in the lab, GAPDH from mouse cells is used. Spectroscopy can also be used to determine and compare make-up of the protein, with techniques including [http://en.wikipedia.org/wiki/UV_spectroscopy UV Spectroscopy], [http://en.wikipedia.org/wiki/Circular_dichroism Circular Dichroism], [http://en.wikipedia.org/wiki/Nuclear_magnetic_resonance_spectroscopy Nuclear Magnetic Resonance] (NMR), and [http://en.wikipedia.org/wiki/Fluorescence Fluorescence]. Further analysis in the lab can be done using [http://en.wikipedia.org/wiki/Western_Blot Western blot]s and [http://en.wikipedia.org/wiki/RT-PCR RT-PCR], but it has to be carefully controlled because under specific conditions it can have various regulation. Visualization and confirmation of important peptides is made possible by using one of a variey of Coomassie staining dyes-- most often [http://en.wikipedia.org/wiki/Coomassie_Brilliant_Blue Coomassie Brilliant Blue] is used for western blots. [http://en.wikipedia.org/wiki/Mass_spectrometry Mass Spectrometry] (MS) and MS/MS methods, used to determine the mass and elemental composition of particles, are also a useful technique to use. It has many variations including [http://en.wikipedia.org/wiki/Fast_atom_bombardment fast atom bombardment] (FAB), [http://en.wikipedia.org/wiki/Thermospray thermospray], and [http://en.wikipedia.org/wiki/Direct_analysis_in_real_time direct analysis in real time] (DART). For more specified experiments concentrating on specific parts of the protein, a variety of GAPDH [http://en.wikipedia.org/wiki/ELISA ELISA] kits have been used which utilize digestive enzymes such as Chymotrypsin and Trypsin to confirm the identity of the peptides. | |||
[[Image:GAPDH western blot.jpg|350 px]] | [[Image:GAPDH western blot.jpg|350 px]] | ||
[[Image:RT-PCR GAPDH.jpg|200 px]] | [[Image:RT-PCR GAPDH.jpg|200 px]] | ||
==References== | ==References== | ||
<references/> | <references/> | ||
#↑Boyer, Paul D.. "The Enzymes - Paul D. Boyer - Google Books." Google Books. N.p., n.d. Web. 1 May 2012. '''<ref>''' Boyer, Paul D.. "The Enzymes - Paul D. Boyer - Google Books." Google Books. N.p., n.d. Web. 1 May 2012. '''[<http://books.google.com/books?id=r6SRFsxYFYwC&pg=PA20&lpg=PA20&dq=GAPDH+cysteine+149&source=bl&ots=0CWwdduzGB&sig=Lb8PtWq7EGdwyvdAnFI_rHYG3tU&hl=en&sa=X&ei=2PyhT-WtHJSy8QTjtsnTCA&ved=0CDMQ6AEwAg#v=onepage&q=GAPDH%20cysteine%20149&f=false>]'''. '''</ref>''' | |||
#↑"GAPDH glyceraldehyde-3-phosphate dehydrogenase [Homo sapiens] - Gene - NCBI." National Center for Biotechnology Information. N.p., n.d. Web. 1 May 2012. '''<ref>'''"GAPDH glyceraldehyde-3-phosphate dehydrogenase [Homo sapiens] - Gene - NCBI." National Center for Biotechnology Information. N.p., n.d. Web. 1 May 2012. '''[<http://www.ncbi.nlm.nih.gov/gene/2597>]'''. '''<ref/>''' | #↑"GAPDH glyceraldehyde-3-phosphate dehydrogenase [Homo sapiens] - Gene - NCBI." National Center for Biotechnology Information. N.p., n.d. Web. 1 May 2012. '''<ref>'''"GAPDH glyceraldehyde-3-phosphate dehydrogenase [Homo sapiens] - Gene - NCBI." National Center for Biotechnology Information. N.p., n.d. Web. 1 May 2012. '''[<http://www.ncbi.nlm.nih.gov/gene/2597>]'''. '''<ref/>''' | ||
#↑"Glyceraldehyde 3-phosphate dehydrogenase." Stanford University. N.p., n.d. Web. 22 Apr. 2012. '''<ref>'''"Glyceraldehyde 3-phosphate dehydrogenase." Stanford University. N.p., n.d. Web. 22 Apr. 2012. '''[<http://www.stanford.edu/~mmogri/bio/gapdh/structure.html>]'''. '''</ref>''' | #↑"Glyceraldehyde 3-phosphate dehydrogenase." Stanford University. N.p., n.d. Web. 22 Apr. 2012. '''<ref>'''"Glyceraldehyde 3-phosphate dehydrogenase." Stanford University. N.p., n.d. Web. 22 Apr. 2012. '''[<http://www.stanford.edu/~mmogri/bio/gapdh/structure.html>]'''. '''</ref>''' | ||
| Line 43: | Line 50: | ||
#↑"Glyceraldehyde 3-phosphate dehydrogenase - Wikipedia, the free encyclopedia." Wikipedia, the free encyclopedia. N.p., n.d. Web. 1 May 2012. '''<ref>'''"Glyceraldehyde 3-phosphate dehydrogenase - Wikipedia, the free encyclopedia." Wikipedia, the free encyclopedia. N.p., n.d. Web. 1 May 2012. '''[<http://en.wikipedia.org/wiki/Glyceraldehyde_3-phosphate_dehydrogenase>]'''. '''</ref>''' | #↑"Glyceraldehyde 3-phosphate dehydrogenase - Wikipedia, the free encyclopedia." Wikipedia, the free encyclopedia. N.p., n.d. Web. 1 May 2012. '''<ref>'''"Glyceraldehyde 3-phosphate dehydrogenase - Wikipedia, the free encyclopedia." Wikipedia, the free encyclopedia. N.p., n.d. Web. 1 May 2012. '''[<http://en.wikipedia.org/wiki/Glyceraldehyde_3-phosphate_dehydrogenase>]'''. '''</ref>''' | ||
#↑"Glyceraldehyde-3-phosphate Dehydrogenase." Biochemistry Dictionary. N.p., n.d. Web. 1 May 2012. '''<ref>'''"Glyceraldehyde-3-phosphate Dehydrogenase." Biochemistry Dictionary. N.p., n.d. Web. 1 May 2012. '''[<http://guweb2.gonzaga.edu/faculty/cronk/biochem/G-index.cfm?definition=GAPDH>]'''. '''</ref>''' | #↑"Glyceraldehyde-3-phosphate Dehydrogenase." Biochemistry Dictionary. N.p., n.d. Web. 1 May 2012. '''<ref>'''"Glyceraldehyde-3-phosphate Dehydrogenase." Biochemistry Dictionary. N.p., n.d. Web. 1 May 2012. '''[<http://guweb2.gonzaga.edu/faculty/cronk/biochem/G-index.cfm?definition=GAPDH>]'''. '''</ref>''' | ||
#↑"Identification of tyrosine nitration in UCH‐L1 and GAPDH - Guingab‐Cagmat - 2011 - ELECTROPHORESIS - Wiley Online Library." Wiley Online Library. N.p., n.d. Web. 1 May 2012. '''<ref>'''"Identification of tyrosine nitration in UCH‐L1 and GAPDH - Guingab‐Cagmat - 2011 - ELECTROPHORESIS - Wiley Online Library." Wiley Online Library. N.p., n.d. Web. 1 May 2012. '''[<http://onlinelibrary.wiley.com/doi/10.1002/elps.201100133/full>]'''. '''</ref>''' | |||
#↑Isupov, M.N., and J.A. Littlechild. "Glyceraldehyde-3-phosphate Dehydrogenase." RCSB Protein Data Bank. N.p., 8 Oct. 1999. Web. 29 Apr. 2012. '''<ref>'''Isupov, M.N., and J.A. Littlechild. "Glyceraldehyde-3-phosphate Dehydrogenase." RCSB Protein Data Bank. N.p., 8 Oct. 1999. Web. 29 Apr. 2012. '''[<www.rcsb.org/pdb/explore/explore.do?structureId=1B7G>]'''. '''</ref>''' | #↑Isupov, M.N., and J.A. Littlechild. "Glyceraldehyde-3-phosphate Dehydrogenase." RCSB Protein Data Bank. N.p., 8 Oct. 1999. Web. 29 Apr. 2012. '''<ref>'''Isupov, M.N., and J.A. Littlechild. "Glyceraldehyde-3-phosphate Dehydrogenase." RCSB Protein Data Bank. N.p., 8 Oct. 1999. Web. 29 Apr. 2012. '''[<www.rcsb.org/pdb/explore/explore.do?structureId=1B7G>]'''. '''</ref>''' | ||
#↑Minter, Melissa. "Glyceraldehyde-3-phosphate Dehydrogenase." Oxidoreductases and the Reactions they Catalyze. N.p., n.d. Web. 20 Apr. 2012. '''<ref>'''Minter, Melissa. "Glyceraldehyde-3-phosphate Dehydrogenase." Oxidoreductases and the Reactions they Catalyze. N.p., n.d. Web. 20 Apr. 2012. '''[<http://www.chem.uwec.edu/Webpapers2005/mintermm/pages/GAPDH.html>]'''. '''</ref>''' | #↑Minter, Melissa. "Glyceraldehyde-3-phosphate Dehydrogenase." Oxidoreductases and the Reactions they Catalyze. N.p., n.d. Web. 20 Apr. 2012. '''<ref>'''Minter, Melissa. "Glyceraldehyde-3-phosphate Dehydrogenase." Oxidoreductases and the Reactions they Catalyze. N.p., n.d. Web. 20 Apr. 2012. '''[<http://www.chem.uwec.edu/Webpapers2005/mintermm/pages/GAPDH.html>]'''. '''</ref>''' | ||
#↑"Structure, Function, and Thermostability of GAPDH." GAPDH. N.p., n.d. Web. 28 Apr. 2012. '''<ref>'''"Structure, Function, and Thermostability of GAPDH." GAPDH. N.p., n.d. Web. 28 Apr. 2012. '''[<www.chem.missouri.edu/TannerGroup/research/gapdh/gapdh.html>]'''.'''</ref>''' | #↑"Structure, Function, and Thermostability of GAPDH." GAPDH. N.p., n.d. Web. 28 Apr. 2012. '''<ref>'''"Structure, Function, and Thermostability of GAPDH." GAPDH. N.p., n.d. Web. 28 Apr. 2012. '''[<www.chem.missouri.edu/TannerGroup/research/gapdh/gapdh.html>]'''.'''</ref>''' | ||
#↑Watson, H.C., and J.C. Campbell. "Twinning In Crystals Of Human Skeletal Muscle D-Glyceraldehyde-3-Phosphate Dehydrogenase." RCSB Protein Data Bank. N.p., 27 Oct. 1983. Web. 28 Apr. 2012. '''<ref>'''Watson, H.C., and J.C. Campbell. "Twinning In Crystals Of Human Skeletal Muscle D-Glyceraldehyde-3-Phosphate Dehydrogenase." RCSB Protein Data Bank. N.p., 27 Oct. 1983. Web. 28 Apr. 2012. '''[<www.rcsb.org/pdb/explore/explore.do?structureId=3gpd>]'''. '''</ref>''' | #↑Watson, H.C., and J.C. Campbell. "Twinning In Crystals Of Human Skeletal Muscle D-Glyceraldehyde-3-Phosphate Dehydrogenase." RCSB Protein Data Bank. N.p., 27 Oct. 1983. Web. 28 Apr. 2012. '''<ref>'''Watson, H.C., and J.C. Campbell. "Twinning In Crystals Of Human Skeletal Muscle D-Glyceraldehyde-3-Phosphate Dehydrogenase." RCSB Protein Data Bank. N.p., 27 Oct. 1983. Web. 28 Apr. 2012. '''[<www.rcsb.org/pdb/explore/explore.do?structureId=3gpd>]'''. '''</ref>''' | ||
Latest revision as of 06:58, 3 May 2012
| This Sandbox is Reserved from 13/03/2012, through 01/06/2012 for use in the course "Proteins and Molecular Mechanisms" taught by Robert B. Rose at the North Carolina State University, Raleigh, NC USA. This reservation includes Sandbox Reserved 451 through Sandbox Reserved 500. | ||||||
To get started:
More help: Help:Editing For more help, look at this link: http://www.proteopedia.org/wiki/index.php/Help:Getting_Started_in_Proteopedia Introduction
|
|
StructureStructure
- While GAPDH possesses four cysteine residues, its is located around , which is responsible for inhibition of glycolysis and fermentation by iodoacetic acid. [1]
- GAPDH has two domains, a GAPDH-like, C-terminal domain, and an NAD Binding Domain.
- The is an Alpha Beta 3-Layer(α-β-α) Sandwich, including amino acids 1-138 and 301-340 on chains O and Q . Its CATH reports a Rossmann fold and a classification as an oxidoreductase.
- The GAPDH-like, C-terminal domain (Catalytic domain) is an Alpha Beta 2-Layer(α-β) Sandwich, including amino acids 139 - 300 on chains O and Q . Its CATH reports a holo-D-Glyceraldehyde-3-Phosphate Dehydrogenase, (domain 2) and a classification as an oxidoreductase (aldehyde(D)-NAD(A)).
- Depending on the environment, including variations in temperature and acidity, GAPDH can have different structural qualities and physical properties. In some cases, it is observed as a twinned structure with an α/β barrel. [2]
ImportanceImportance
Role in Glycolysis:Role in Glycolysis:
The Steps:The Steps:
- In two coupled steps,
GAPDH catalyzes the conversion of glyceraldyhyde-3-phosphate at carbon 1 to 1,3-bisphosphoglycerate (1,3-BPG).
The Reactions:The Reactions:
- The reaction involving the GAPDH enzyme combines phosphorylation with oxidation in an overall endergonic reaction. (ΔG°'=+6.3 kJ/mol (+1.5 kcal/mol)) First, the oxidation of glyceraldyhyde-3-phosphate to D-glycerate 1,3-bisphosphate takes place, in which an aldehyde is converted to carboxylic acid ((ΔG°'=-50 kJ/mol (-12 kcal/mol))and NAD+ (Nicotinamide adenine dinucleotide), an important co-factor and ligand found bound to the of GAPDH, is simultaneously reduced endergonically to NADH. This oxidation reaction is required for the initiation of the second reaction because it is highly exergonic and thus drives the endergonic second reaction ((ΔG°'=+50 kJ/mol (+12 kcal/mol)). In the second reaction a molecule of inorganic phosphate is transferred to a GAP intermediate to form a product with a high potential to transfer phosphates, 1,3-bisphosphoglycerate.
The Mechanism:The Mechanism:
- The mechanism of GAPDH is mainly dependent the thiol functional group on a cysteine residue, of the 3-phosphoglycerate amino acid biosynthetic family, which lies within the . This important thiol group acts as a nucleophile, attacking the carbon on the aldehyde functional group of the substrate, glyceraldehyde 3-phosphate, after it binds to GAPDH. The creation of a thiohemiacetal intermediate occurs from this oxidative reaction, which then loses a hydride to NAD+, which is also bound nearby, forming NADH and a carboxyl group from the aldehyde. Conservation of energy is maintained through this thioester linkage to the active site cysteine. The GAP intermediate is then used in the second step, where it is important to notice that without GAPDH's use of covalent catalysis, the energy barrier of the reaction would be too high and the reaction would be too slow for living organisms.
Other roles:Other roles:
- The importance of GAPDH in transcription was discovered by Zheng et. al. in 2003. It was found that OCA's transcriptional coactivator complex contains GAPDH, which moves between the cytosol and nucleus and may link the metabolic state to gene transcription.
- In 2005 the initiation of apoptosis was shown to be mediated by GAPDH by Hara et. al. when it was found to bind to DNA like it does in transcription activation.
GAPDH was also found to be involved in ER to Golgi transport because it is recruited by rab2 to vesicular-tubular clusters of the endoplasmic reticulum where it helps form COP 1 vesicles.
- It was also found that GAPDH is plays a role in certain neruodegenerative disorders, as is able to find stretches which are encoded by the gene's CAG repeats and bind to the gene products formed from disorders such as Huntington's disease, Alzheimer’s disease, Parkinson’s disease and Machado-Joseph disease.
Use in the lab:Use in the lab:
Quantitation:Quantitation:
- Overall, GAPDH is one of the most common “housekeeping genes” and is found in high levels in tissues and cells so it is commonly used in biological research as a loading control in quantification techniques such as Lowry Assays, Bradford Assays, Isoelectric Focusing, 2D-PAGE, Size Exclusion Chromatography (SEC), as well as 1-D and 2-D SDS-PAGE when it is conjugated to a known amount of biotin or BODIPY fluorophore to create a standard. A good method of quantitation lies under proteomics, which concompasses a variation of techniques including Differential in-gel Electrophoresis (DIGE), SILAC (for Metabolic Labeling), iTRAQ (for Chemical Labeling).
Qualification and Analysis:Qualification and Analysis:
- Analysis of GAPDH can give much useful information when comparing proteins and substances in an experiment, thus some techniques used include Affinity Chromatography, Ion Exchange Chromatography, Reversed Phase LC. Often times in the lab, GAPDH from mouse cells is used. Spectroscopy can also be used to determine and compare make-up of the protein, with techniques including UV Spectroscopy, Circular Dichroism, Nuclear Magnetic Resonance (NMR), and Fluorescence. Further analysis in the lab can be done using Western blots and RT-PCR, but it has to be carefully controlled because under specific conditions it can have various regulation. Visualization and confirmation of important peptides is made possible by using one of a variey of Coomassie staining dyes-- most often Coomassie Brilliant Blue is used for western blots. Mass Spectrometry (MS) and MS/MS methods, used to determine the mass and elemental composition of particles, are also a useful technique to use. It has many variations including fast atom bombardment (FAB), thermospray, and direct analysis in real time (DART). For more specified experiments concentrating on specific parts of the protein, a variety of GAPDH ELISA kits have been used which utilize digestive enzymes such as Chymotrypsin and Trypsin to confirm the identity of the peptides.
ReferencesReferences
- ↑ Boyer, Paul D.. "The Enzymes - Paul D. Boyer - Google Books." Google Books. N.p., n.d. Web. 1 May 2012. [<http://books.google.com/books?id=r6SRFsxYFYwC&pg=PA20&lpg=PA20&dq=GAPDH+cysteine+149&source=bl&ots=0CWwdduzGB&sig=Lb8PtWq7EGdwyvdAnFI_rHYG3tU&hl=en&sa=X&ei=2PyhT-WtHJSy8QTjtsnTCA&ved=0CDMQ6AEwAg#v=onepage&q=GAPDH%20cysteine%20149&f=false>].
- ↑ Watson, H.C., and J.C. Campbell. "Twinning In Crystals Of Human Skeletal Muscle D-Glyceraldehyde-3-Phosphate Dehydrogenase." RCSB Protein Data Bank. N.p., 27 Oct. 1983. Web. 28 Apr. 2012. [<www.rcsb.org/pdb/explore/explore.do?structureId=3gpd>].
- ↑Boyer, Paul D.. "The Enzymes - Paul D. Boyer - Google Books." Google Books. N.p., n.d. Web. 1 May 2012. [1]
- ↑"GAPDH glyceraldehyde-3-phosphate dehydrogenase [Homo sapiens] - Gene - NCBI." National Center for Biotechnology Information. N.p., n.d. Web. 1 May 2012. Cite error: Closing
</ref>missing for<ref>tag - ↑"Glyceraldehyde 3-phosphate dehydrogenase (EC 1." European Bioinformatics Institute | Homepage | EBI. N.p., n.d. Web. 1 May 2012. [2]
- ↑"Glyceraldehyde 3-phosphate dehydrogenase - Wikipedia, the free encyclopedia." Wikipedia, the free encyclopedia. N.p., n.d. Web. 1 May 2012. [3]
- ↑"Glyceraldehyde-3-phosphate Dehydrogenase." Biochemistry Dictionary. N.p., n.d. Web. 1 May 2012. [4]
- ↑"Identification of tyrosine nitration in UCH‐L1 and GAPDH - Guingab‐Cagmat - 2011 - ELECTROPHORESIS - Wiley Online Library." Wiley Online Library. N.p., n.d. Web. 1 May 2012. [5]
- ↑Isupov, M.N., and J.A. Littlechild. "Glyceraldehyde-3-phosphate Dehydrogenase." RCSB Protein Data Bank. N.p., 8 Oct. 1999. Web. 29 Apr. 2012. [6]
- ↑Minter, Melissa. "Glyceraldehyde-3-phosphate Dehydrogenase." Oxidoreductases and the Reactions they Catalyze. N.p., n.d. Web. 20 Apr. 2012. [7]
- ↑"Structure, Function, and Thermostability of GAPDH." GAPDH. N.p., n.d. Web. 28 Apr. 2012. [8]
- ↑Watson, H.C., and J.C. Campbell. "Twinning In Crystals Of Human Skeletal Muscle D-Glyceraldehyde-3-Phosphate Dehydrogenase." RCSB Protein Data Bank. N.p., 27 Oct. 1983. Web. 28 Apr. 2012. [9]