User:Mitchell Long/Sandbox 1
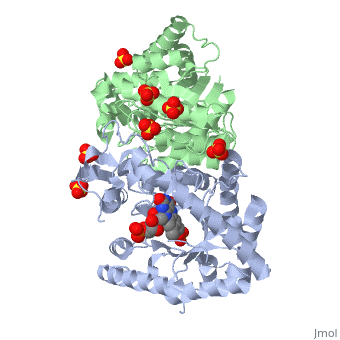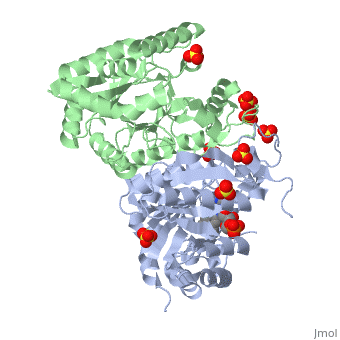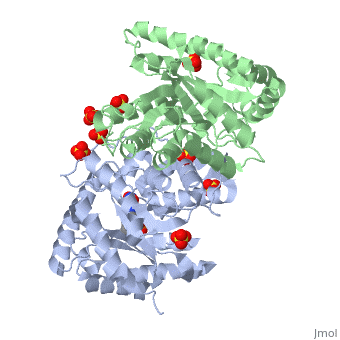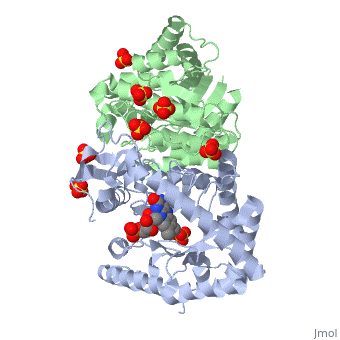
IntroductionLuciferases are a class of enzymes that catalyze the oxidation of a long chain aliphatic aldehydes the emission of blue-green light. The luciferase found in 'Vibrio harveyi' is a heterodimer that is composed of a catalytic α subunit and a homologous but noncatalytic β subunit. This reaction results in the formation of a carboxylic acid, reduced flavinmononucleotide and the emission of photons in the form of blue-green light. The catalytic α subunit houses the active site and is connected to the β subunit via a single interatcion between the mobile loop and the α subunit at α Phe 272 and Tyr 151 of the β subunit.
MechanismLuciferase found in'V. Harveyi' binds noncovalently to a reduced flavin mononucleotide cofactor, an aliphatic aldehyde and oxygen to yield oxidized flavin mononucleotide, water, and carboxylic acid. The reaction occurs in two steps forming a hydroxyflavin intermediate and ultimately results in the oxidation of the aldehyde and emission of photonsCite error: Invalid parameter in FMNH2+O2+RCHO→FMN+RCOOH+H2O+hv(490nm) The catalytic α subunit houses the FMN cofactor and is connected to the β subunit via a hairpin structure called the "." The organic substrate for bacterial luciferase in vivo is myristic aldehyde, although many aliphatic aldehydes of various lengths can induce bioluminescence in vitroCite error: Invalid parameter in
Structural MotifsStructure homology-There is a great deal of sequence homology and structural coservation between the α and β subunits. When superimposed over the barrels of the alpha and beta subunits with a deviation of 0.62Å for 42 equivalent α carbons. The region of the beta subunit that contains the 29 residue deletion with respect to the alpha subunit differs notably in arrangementCite error: Invalid parameter in Active Site and Alpha Subunit-the of bacterial luciferase is a large open cavity that is accessible to solvent via an opening located at the C-terminal ends of the ǰ strands of the TIM-barrel structureCite error: Invalid parameter in
The β subunit-The beta subunit is characterized as a necessary but non-catalytic subunit that stabilizes the catalytic α subunit that is responsible for the oxidation reaction. The beta and alpha subunits are connected by a single interaction between the
Mobile Loop- Residues 272-288 on the α are known as the mobile loop. This portion of the alpha subunit contains a single residue that forms a salt bridge with the beta subunit and stabilizes the active siteCite error: Invalid parameter in
(β/α)8 TIM Barrel- The tertiary structure of the α and β subunits is very similar. While both the alpha and beta subunits are similar, the alpha subunit contains an extra 29 residues that the beta lacks. Both subunits fold into a single-domain eight-stranded β/α barrel motif. the two subunits assemble around a parallel four-helix bundle centered on a pseudo 2-fold axis that relates the alpha and beta subunitsCite error: Invalid parameter in |
| ||||||||||
| |||||||||
| 3fgc, resolution 2.30Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , , | ||||||||
| Gene: | luxA (Vibrio harveyi), luxB (Vibrio harveyi) | ||||||||
| Activity: | Alkanal monooxygenase (FMN-linked), with EC number 1.14.14.3 | ||||||||
| Related: | 1luc, 1brl | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Applications In BiotechnologyApplications In Biotechnology
Luciferases are most commonly used as reporter genes by transduction or transfection assays. Reporter genes are inserted into an organism with a gene of interest. This is a powerful method of measuring gene expression because it is non-invasive. Genes for luciferase can paired with an inducible operon. When the gene for luciferase and the gene of interest are incorporated into the host genome, they can "turned on" by induction. Once the desired gene is turned on, gene expression can be determined by the intensity of the light produced by transcription of the gene of interest.
Quorum SensingQuorum Sensing
In a process known as quorum sensing, bacteria communicate using secreted signal molecules called autoinducers(AIs). 'V. harveyi' is a mesophilic, gram negative, rod shaped bacteria that can communicate with other bacteria via quorum sensing. Quorum-sensing bacteria alter gene expression in response to the accumulation of AIs, which reflects an increase in cell population densityCite error: Invalid parameter in <ref> tag. This process is believed to provide bacteria a means to coordinately control the gene expression of the group, giving them multicellular characteristics. When bacteria reach a "quorum," their population has reached a density high enough to coordinate gene expressionCite error: Invalid parameter in <ref> tag. Often, bacteria make and respond to multiple AIs. Vibrio harveyi, a free-living marine bacterium, produces at least three distinct AIs to control bioluminescence, biofilm formation, Type III Secretion (TTS), and protease production. When a bacterial population density is low, the LuxI gene is transcribed constitutively at basal level. The three V. harveyi AIs are HAI-1, an acyl homoserine lactone; AI-2, a furanosyl-borate-diester; and CAI-1, of unknown structureCite error: Invalid parameter in <ref> tag. When the population density reaches an adequate level, the conjugate receptor LuxR begins transcription. LuxR is the regulatory receptor, and when an AI binds the the LuxR receptor, transcription is turned on resulting in the production of more AI and the expression of other genes involved in quorum sensing. When V. harveyi reaches a high enough population density, it's quorum sensing genes are activated and the transcription of the genes that code for the luciferase enzyme.
Cite error: Invalid parameter in <ref> tag
Cite error: Invalid parameter in <ref> tag
Cite error: Invalid parameter in <ref> tag