To get started:
- Click the edit this page tab at the top. Save the page after each step, then edit it again.
- Click the 3D button (when editing, above the wikitext box) to insert Jmol.
- show the Scene authoring tools, create a molecular scene, and save it. Copy the green link into the page.
- Add a description of your scene. Use the buttons above the wikitext box for bold, italics, links, headlines, etc.
More help: Help:Editing
For more help, look at this link:
http://www.proteopedia.org/wiki/index.php/Help:Getting_Started_in_Proteopedia
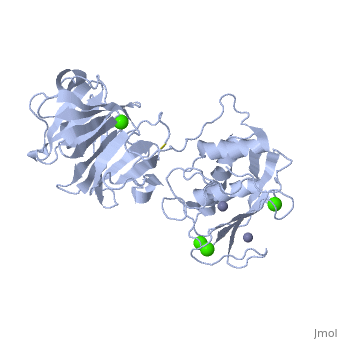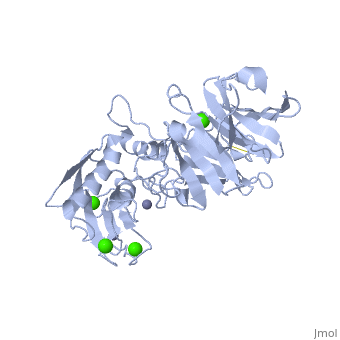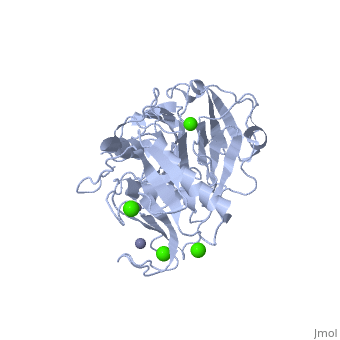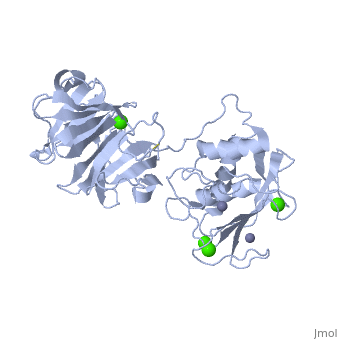
Matrix Metalloproteinase 1(MMP1) also known as Interstitial collagenase is a zinc-dependent protease that degrades extracellular matrix proteins.[1] Collagenases are enzymes, which cleave bonds in collagen. MMP1 belongs to the Matrix metalloproteinase (MMP) family, which are involved in the regulation of cell-matrix composition by the breakdown of the extracellular matrix in normal physiological processes. These physiological processes many include disease activities such as arthritis and metastasis as well as normal human reproduction and embryonic development.[2]
|
|
| 2clt, resolution 2.67Å ()
|
| Ligands:
|
,
|
| Activity:
|
Interstitial collagenase, with EC number 3.4.24.7
|
| Related:
|
1ayk, 1cge, 1cgf, 1cgl, 1hfc, 1su3, 2ayk, 2tcl, 3ayk, 4ayk, 966c
|
|
|
|
|
|
|
| Resources:
|
FirstGlance, OCA, RCSB, PDBsum
|
| Coordinates:
|
save as pdb, mmCIF, xml
|
StructureStructure
The Structure of MMP-1 and all other members of the Metalloproteinase family for that matter are formed from three domains. The structure comprises of the N-terminal catalytic domain, the linker region and the C-terminal hemopexin domain. The structure of human MMP-1 was determined with X-Ray Crystallography at a resolution of 2.67A to have two monomers (chains A and B).[3] The catalytic domain of one monomer contacts the hemopexin domain of the other monomer. An interesting observation that has been noted is that the contact site used by the two monomers in the asymmetric unit to form the dimer is not the same as the dimerization site observed in the structure of the MMP-9 hemopexin domain. This difference shows that not all members of the Matrix Metalloproteinase family behave the same in their dimerization processes. Another interesting feature about the protein is that can be found within all areas of the protein rather than being located near a certain domain.
Catalytic DomainCatalytic Domain
Many of the MMPs share similar catalytic domain structures and this holds up for MMP-1. is about 160 amino acid residues in length with the catalytic zinc ion residing in the C-terminal segment of this domain. The Domain consists of three α-helices, a highly twisted five-stranded β-sheet, and three calcium-binding sites, which are used for ligand binding. Except for one calcium that is held within monomer A, all of the calcium ions are coordinated with either four or five liganding residues.
Linker RegionLinker Region
The Catalytic Domain is followed by a Linker region spanning 15-65 residues in all MMPs.[4] The linker region connects the Catalytic Domain and the Hemopexin Domain of the enzyme. This leads to the MMP-1 being more stabilized overall, which is highly required for the mutual actions of two domains.
Hemopexin-like DomainHemopexin-like Domain
The consists of about 210 amino acids and is composed of four Hemopexin modules (I-IV), each representing a blade of the beta-propeller structure. Each blade starts with either the DAA or DAX motif, in which Asp residues direct the central calcium ion through their carbonyl oxygen atom. [5] [3] Glu310 also provides the fourth coordination thus completing the acidic patch at the entrance of the central, solvent-accessible channel. The side chains of these residues also help to form salt bridges with β-strands and hold the entrance of the central channel inline.
Three molecules of water are held within the center of the central channel and they are not involved in the geometry of the calcium ion at the center of the tunnel. It has been reasoned that the presence of these ions is not related to the stability conditions but rather a consequence of the crystallization methods.
Mechanism of ActionMechanism of Action
Several Studies have been conducted on the specific mechanism of MMPs, and many different perspectives of the mechanism have been published. The difference in the proposed mechanisms shows that the research within this area is still undergoing and there is no final verdict as to the mechanism of action. Robert Visse et al, states in a review of Matrix Metalloproteinases, that MMPs can be activated by proteinases or in vitro by chemical agents, such as thiol-modifying agents (4-aminophenylmercuric acetate, HgCl2, and N-ethylmaleimide), oxidized glutathione, SDS, chaotropic agents, and reactive oxygens. [2] These agents mostly work to disturb the interaction between the cysteine-zinc of the cysteine switch. Proteolytic activation of MMPs is stepwise with the initial proteolytic attack occuring at an exposed loop region between the first and the second helices of the propeptide, once a portion of the propetide is removed, then the rest of the propeptide is destabilized which allows for the intermolecular processing by partially activated MMP intermediates or other MMPs. The final step in the activation process is therefore conducted by an MMP
This image is a representation of the mode of activation that is utilized in MMP-1. the image is used to show activation in human colleganase (proMMP-1)[2] 
Medical ImplicationsMedical Implications
MMP-1 along with all the other MMPs are a family of zinc-binding endopeptidases that degrade most of the components of the extracellular matrix (ECM). Their influence within the medical community has mainly come from the human tumor cells. MMPs have been frequently found to be overexpressed in most forms of human tumors. Scientists have determined that based on their degrading activity. they are believed to be to contribute to the proliferation, invasion, and metastasis of tumor cells by eliminating the surrounding ECM barrier. MMPs are required for tumor-induced angiogenesis, and so inhibition of the MMPs would be expected to stabilize malignant growth. MMP inhibitors have been developed to counteract these effects however they have been seen as ineffective against human tumors. Beyond their role in tumors MMPs have also bee seen to have a role in regulation of the ECM. MMPs promote cell interaction and migration as well as degradation by processing cell adhesion molecules such as CD44 and integrin αv chain. [6]
ReferencesReferences
- ↑ Nagase, H., Barrett, A. J., Woessner, J. F., Jr. Nomenclature and glossary of the matrix metalloproteinases. Matrix Suppl. 1: 421-424, 1992.
- ↑ 2.0 2.1 2.2 Visse, Robert, and Hideaki Nagase. "Matrix Metalloproteinases and Tissue Inhibitors of Metalloproteinases." Circulation Research 92 (2003): 827-39.
- ↑ 3.0 3.1 Shalini Iyer, Robert Visse, Hideaki Nagase, K. Ravi Acharya, Crystal Structure of an Active Form of Human MMP-1, Journal of Molecular Biology, Volume 362, Issue 1, 8 September 2006, Pages 78-88, ISSN 0022-2836, 10.1016/j.jmb.2006.06.079.
- ↑ "2.8. Matrix Metalloproteinases (MMPs)." Matrix Metalloproteinases (MMPs). Web. 02 May 2012. <http://herkules.oulu.fi/isbn9514270789/html/x561.html>.
- ↑ J Li, P Brick, MC O'Hare, T Skarzynski, LF Lloyd, VA Curry, IM Clark, HF Bigg, BL Hazleman, TE Cawston, DM Blow, Structure of full-length porcine synovial collagenase reveals a C-terminal domain containing a calcium-linked, four-bladed β-propeller, Structure, Volume 3, Issue 6, June 1995, Pages 541-549
- ↑ Arnold LH, Butt LE, Prior SH, Read CM, Fields GB, Pickford AR. The interface between catalytic and hemopexin domains in matrix metalloproteinase-1 conceals a collagen binding exosite. J Biol Chem. 2011 Dec 30;286(52):45073-82. Epub 2011 Oct 26. PMID:22030392 doi:10.1074/jbc.M111.285213
|