Histone
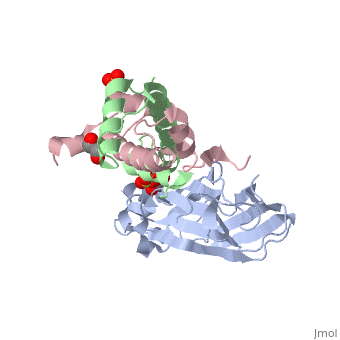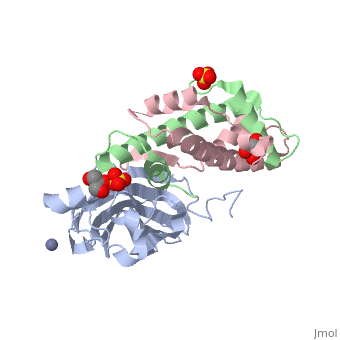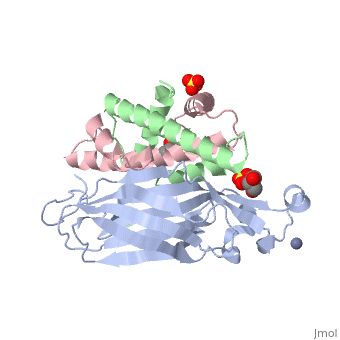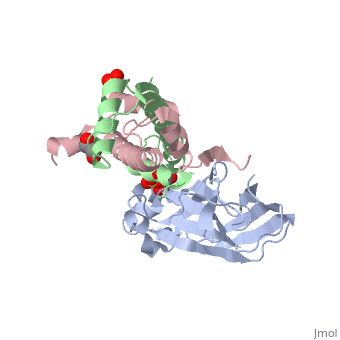
Histone core protein structureHistones are highly (more purple = more conserved) with (blue is positive charge, red is negative charge). Because of this positive charge, they interact electrostatically with the negatively charged phosphate groups in DNA. There are five major classes of histones: H1/H5, H2A, H2B, H3, and H4.[1][2] Histones , , , and are known as the core histones, while histones H1 and H5 are known as the linker histones. The 4 'core' histones (H2A, H2B, H3 and H4) are relatively similar in structure and are highly conserved through evolution, all featuring a motif (which allows the easy dimerization). They also share the feature of long 'tails' on one end of the amino acid structure, which are often covalently modified to regulate gene expression.
Histone interactions with DNAHistones are the chief protein components of , acting as spools around which DNA winds, and play a role in gene regulation. Without histones, the unwound DNA in chromosomes would be very long; each human cell has about 1.8 meters of DNA, but wound on the histones it has about 90 micrometers (0.09 mm) of chromatin, which, when duplicated and condensed during mitosis, result in about 120 micrometers of chromosomes.[3] DNA is wrapped around nucleosomes with approximately 50 base pairs of DNA between subsequent nucleosomes (also referred to as linker DNA). The assembled histones and DNA is called chromatin. During mitosis and meiosis, the condensed chromosomes are assembled through interactions between nucleosomes and other regulatory proteins.
The nucleosome core is formed of two and a , forming two nearly by tertiary structure.[4] 147 base pairs of around this core particle 1.65 times in a left-handed super-helical turn.[4] The linker histone H1 binds the nucleosome and the entry and exit sites of the DNA, thus locking the DNA into place[5] and allowing the formation of higher order structure. The chromatosome contains histone H1 binding to nucleosome. It contains 166 DNA base pairs.
In all, histones make five types of interactions with DNA:
In general, genes that are active have less bound histone, while inactive genes are highly associated with histones during interphase. It also appears that the structure of histones has been evolutionarily conserved, as any deleterious mutations would be severely maladaptive. Chromatin regulationHistones are subject to post translational modification by enzymes primarily on their N-terminal tails, but also in their globular domains. Such modifications include methylation, acetylation, phosphorylation, SUMOylation, ubiquitination, and ADP-ribosylation. This affects gene expression. The core of the histones H2A, H2B, and H3 can also be modified. Combinations of modifications are thought to constitute a code, the so-called "histone code".[6][7] Histone modifications act in diverse biological processes such as gene regulation, DNA repair, chromosome condensation (in mitosis, spermatogenesis, and meiosis).[8] The common nomenclature of histone modifications is:
So H3K4me1 denotes the monomethylation of the 4th residue (a lysine) from the start (i.e., the N-terminal) of the H3 protein.
|
| ||||||||||
- For nucleosome structure see
- User:Eric Martz/Nucleosomes
- Nucleosome structure
- Nucleosomes
- Nucleosome structure (Spanish)
3D Structures of histone3D Structures of histone
ReferencesReferences
- ↑ Bhasin M, Reinherz EL, Reche PA. Recognition and classification of histones using support vector machine. J Comput Biol. 2006 Jan-Feb;13(1):102-12. doi: 10.1089/cmb.2006.13.102. PMID:16472024 doi:http://dx.doi.org/10.1089/cmb.2006.13.102
- ↑ Template:Cite book
- ↑ Redon C, Pilch D, Rogakou E, Sedelnikova O, Newrock K, Bonner W. Histone H2A variants H2AX and H2AZ. Curr Opin Genet Dev. 2002 Apr;12(2):162-9. doi: 10.1016/s0959-437x(02)00282-4. PMID:11893489 doi:http://dx.doi.org/10.1016/s0959-437x(02)00282-4
- ↑ 4.0 4.1 Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997 Sep 18;389(6648):251-60. PMID:9305837 doi:10.1038/38444
- ↑ Template:Cite book
- ↑ Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000 Jan 6;403(6765):41-5. doi: 10.1038/47412. PMID:10638745 doi:http://dx.doi.org/10.1038/47412
- ↑ Jenuwein T, Allis CD. Translating the histone code. Science. 2001 Aug 10;293(5532):1074-80. doi: 10.1126/science.1063127. PMID:11498575 doi:http://dx.doi.org/10.1126/science.1063127
- ↑ Song N, Liu J, An S, Nishino T, Hishikawa Y, Koji T. Immunohistochemical Analysis of Histone H3 Modifications in Germ Cells during Mouse Spermatogenesis. Acta Histochem Cytochem. 2011 Aug 27;44(4):183-90. doi: 10.1267/ahc.11027. Epub, 2011 Jul 20. PMID:21927517 doi:http://dx.doi.org/10.1267/ahc.11027