Escherichia coli LepA, the ribosomal back translocase
<StructureSection load= size='350' side='right' scene='3cb4/3cb4biolunit/1' caption='GTP-binding protein LepA (PDB code 3cb4)'>
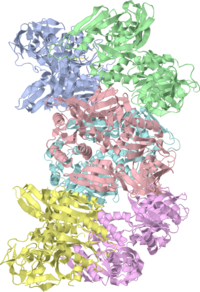
The Crystal Structure of LepA, the ribosomal back translocaseThe Crystal Structure of LepA, the ribosomal back translocase
LepA is a highly conserved elongation factor that promotes the back translocation of tRNAs on the ribosome during the elongation cycle. We have determined the crystal structure of LepA from Escherichia coli at 2.8-A resolution. The high degree of sequence identity between LepA and EF-G is reflected in the structural similarity between the individual homologous domains of LepA and EF-G. However, the orientation of domains III and V in LepA differs from their orientations in EF-G. LepA also contains a C-terminal domain (CTD) not found in EF-G that has a previously unobserved protein fold. The high structural similarity between LepA and EF-G enabled us to derive a homology model for LepA bound to the ribosome using a 7.3-A cryo-EM structure of a complex between EF-G and the 70S ribosome. In this model, the very electrostatically positive CTD of LepA is placed in the direct vicinity of the A site of the large ribosomal subunit, suggesting a possible interaction between the CTD and the back translocated tRNA or 23S rRNA.
The structure of LepA, the ribosomal back translocase., Evans RN, Blaha G, Bailey S, Steitz TA, Proc Natl Acad Sci U S A. 2008 Mar 25;105(12):4673-8. Epub 2008 Mar 24. PMID:18362332
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.
About this StructureAbout this Structure
The BasicsThe Basics
The
LepA and EF-G Are SimilarLepA and EF-G Are Similar
The domains are colored according to figure 1 of the paper describing the structure.
Note that the individual domains of LepA and EF-G, such as III V, superimpose much better when done separately (shown in figure 2 of the paper describing the structure).
PDB EntryPDB Entry
3cb4 is a 6 chains structure of sequences from Escherichia coli k12. Full crystallographic information is available from OCA.
Related Structures and TopicsRelated Structures and Topics
- Ef-G
- The Ribosome
- 70S ribosome bound to EF-G: 4v5f
- For additional information, see: Translation
3D structures of LepA3D structures of LepA
Updated on 28-December-2017
3cbd – EcLepA – Escherichia coli
2ywe – AaLepA – Aquifex aeolicus
2ywf – AaLepA + GMPPNP
2ywg – AaLepA + GTP
2ywh – AaLepA + GDP
3deg – EcLepA + 70S ribosome - CryoEM
4k8w – LepA arm-swapped dimer – Streptococcus pyogenes
ReferenceReference
Page begun with original page seeded for PDB entry 3cb4 by OCA on Wed Jun 3 08:45:42 2009