Atragin
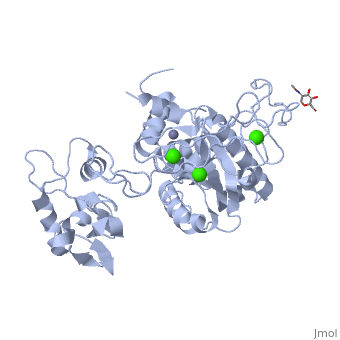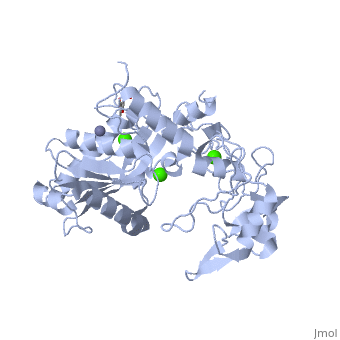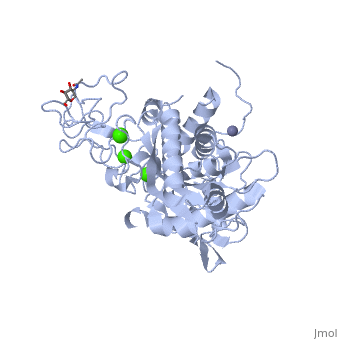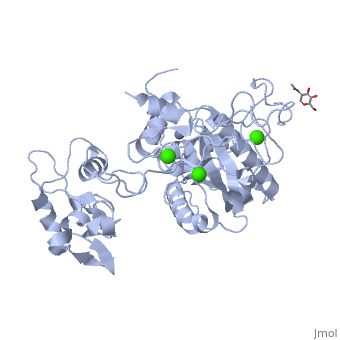
IntroductionAtragin (MW 49.7 kDa and theoretical pI 8.6) is a Snake Venom MetalloProtease (SVMP) belonging to the ADAM/adamlysin/ reprolysin family and is found in the venom of the Naja Atra, or Chinese cobra [1]. It contains a metalloprotease domain (M),disintegrin-like domain(D), and cysteine-rich domain (C), making it a P-III SVMP [2]. When injected into the bloodstream, P-III SVMPs cause hemorrhagic effects, inhibit platelet formation and inhibit cell migration activity[1][3]. These P-III SVMPs bear very close resemblance to mammalian ADAM (A Disintegrin And Metalloprotease) proteins, which also contain the MDC domain architecture [4]. The ADAMs are a group of around 40 transmembrane proteins that have been found so far in mammals, 19 of which genes are found in humans [5][6]. These proteins play a role in the production of cytokines and growth factors, such as ADAM 17 which releases tumor necrosis factor alpha (TNFa) as an immunomodulatory and pro-inflammatory cytokine [7]; and ADAM 9 which has been shown to be involved in the release of heparin-binding EGF, which inhibits the proliferation of neighboring cells [8]. ADAM proteins have also been shown to be involved with multiple human diseases today, including cancer, asthma, cardiac hypertrophy and SARS [9][10][11][12]. Unfortunately, the full structure of the ADAM proteins is not yet available and our current understanding of the ADAM structure is based primarily on viper P-III SVMPs[4][13][14][14][15] . Atragin, however, is an elapid P-III SVMP and provides more clues as to how the MDC domains work together. Structureis a 63x52x67 Å protein that contains a Metalloprotease domain, Disintegrin-like domain, and a cysteine-rich domain[1]. The overall architecture of the protein gives a structure that is similar to other P-III SVMPs such as VAP1 [14] in that the recognition site of the C domain faces the M domain’s catalytic cleft in a linear orientation, which means the target recognized by the C domain could be the same as the substrate catalyzed by M domain. This is a contrast to other P-III SVMPs, such as the K-like protein shown below, which can have an I-shaped structure that contains a non-linear orientation between the C and M domains, in which the target recognized by the C domain can be different than the catalyzed substrate molecule [1]. 'Metalloprotease Domain The Metalloprotease domain (M) consists of and is a metalloendopeptidase. Every endometalloprotease contains a strain of residues needed for zinc and substrate binding for proteolysis. In Atragin, the binding sequence can be found from aa 341-351. The atom is ligated by the side chains of His341 and 345 on the a-helix, and this allows His 351 at the turn the be responsible for the catalytic reaction [16]. One of the ways that the N. atra is able to prevent self-proteolysis is by storing the atragrin protein in a venom lumen gland at an acidic pH and containing citrate and the tripeptide pyroglutamyl-lysyl-tryptophan enzymatic inhibitor in the gland[1][17][18]. In acidic conditions, there is no detectable electron density at the flexible loop, which contains the Met-turn. Under acidic conditions, the prononated histidines cause a structural change which results in an unfixed position of the zinc atom [1]. The picture below shows the general movement of the histidines. 'Disintegrin-like Domain Following the M domain and a linker , is the (D) domain. This domain is thought to play an important role in the relative orientation of the M and C domains in P-III SVMPs (Think back to C-shaped versus I-shaped). Atragin maintains the C-shaped architecture as its domain has three disulfide bonds, and its conatins another 3 disulfide bonds, which is similar to other C-shaped proteins [4][14]. These multiple disulfide bonds make the protein structure very stable. One also connects these two subdomains. The disulfide bond pattern in the D domain alters the orientations of the in the ADAM/adamalysin/ reprolysins family [1]. The different orientations could explain some of the ADAM enzymatic processes, seeing as the different lengths or altering the disulfide pairs of the D-domain can increase or decrease the size of the cleft and the orientation. This could cause for substrates of different sizes and shapes to able to be cleaved by the M-domain in ADAMs [1]. 'Cysteine-rich Domain The Cysteine-rich (C) domain contains the domains. The C-hand subdomain is the most intriguing to most researches because it contains the Hyper Variable Region (HVR. This region is through to play an important role in target selection since it sits on the inside of the C-shaped structures cleft [14]. The C domain of Atragin consists of seven , and this suggests it may play a functional role. The HVR of Atragin is also though to be responsible for the inhibitory affect on cell migration activity since it is exposed to target recognition [1]. |
| ||||||||||
3D structures of atragin3D structures of atragin
ReferencesReferences
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 Guan HH, Goh KS, Davamani F, Wu PL, Huang YW, Jeyakanthan J, Wu WG, Chen CJ. Structures of two elapid snake venom metalloproteases with distinct activities highlight the disulfide patterns in the D domain of ADAMalysin family proteins. J Struct Biol. 2010 Mar;169(3):294-303. Epub 2009 Nov 22. PMID:19932752 doi:10.1016/j.jsb.2009.11.009
- ↑ Fox JW, Serrano SM. Structural considerations of the snake venom metalloproteinases, key members of the M12 reprolysin family of metalloproteinases. Toxicon. 2005 Jun 15;45(8):969-85. Epub 2005 Apr 9. PMID:15922769 doi:10.1016/j.toxicon.2005.02.012
- ↑ Gutierrez JM, Rucavado A. Snake venom metalloproteinases: their role in the pathogenesis of local tissue damage. Biochimie. 2000 Sep-Oct;82(9-10):841-50. PMID:11086214
- ↑ 4.0 4.1 4.2 Igarashi T, Araki S, Mori H, Takeda S. Crystal structures of catrocollastatin/VAP2B reveal a dynamic, modular architecture of ADAM/adamalysin/reprolysin family proteins. FEBS Lett. 2007 May 29;581(13):2416-22. Epub 2007 Apr 30. PMID:17485084 doi:10.1016/j.febslet.2007.04.057
- ↑ Seals DF, Courtneidge SA. The ADAMs family of metalloproteases: multidomain proteins with multiple functions. Genes Dev. 2003 Jan 1;17(1):7-30. PMID:12514095 doi:10.1101/gad.1039703
- ↑ White JM. ADAMs: modulators of cell-cell and cell-matrix interactions. Curr Opin Cell Biol. 2003 Oct;15(5):598-606. PMID:14519395
- ↑ Moss ML, Jin SL, Milla ME, Bickett DM, Burkhart W, Carter HL, Chen WJ, Clay WC, Didsbury JR, Hassler D, Hoffman CR, Kost TA, Lambert MH, Leesnitzer MA, McCauley P, McGeehan G, Mitchell J, Moyer M, Pahel G, Rocque W, Overton LK, Schoenen F, Seaton T, Su JL, Becherer JD, et al.. Cloning of a disintegrin metalloproteinase that processes precursor tumour-necrosis factor-alpha. Nature. 1997 Feb 20;385(6618):733-6. PMID:9034191 doi:10.1038/385733a0
- ↑ Iwamoto R, Yamazaki S, Asakura M, Takashima S, Hasuwa H, Miyado K, Adachi S, Kitakaze M, Hashimoto K, Raab G, Nanba D, Higashiyama S, Hori M, Klagsbrun M, Mekada E. Heparin-binding EGF-like growth factor and ErbB signaling is essential for heart function. Proc Natl Acad Sci U S A. 2003 Mar 18;100(6):3221-6. Epub 2003 Mar 5. PMID:12621152 doi:10.1073/pnas.0537588100
- ↑ Asakura M, Kitakaze M, Takashima S, Liao Y, Ishikura F, Yoshinaka T, Ohmoto H, Node K, Yoshino K, Ishiguro H, Asanuma H, Sanada S, Matsumura Y, Takeda H, Beppu S, Tada M, Hori M, Higashiyama S. Cardiac hypertrophy is inhibited by antagonism of ADAM12 processing of HB-EGF: metalloproteinase inhibitors as a new therapy. Nat Med. 2002 Jan;8(1):35-40. PMID:11786904 doi:10.1038/nm0102-35
- ↑ Haga S, Yamamoto N, Nakai-Murakami C, Osawa Y, Tokunaga K, Sata T, Yamamoto N, Sasazuki T, Ishizaka Y. Modulation of TNF-alpha-converting enzyme by the spike protein of SARS-CoV and ACE2 induces TNF-alpha production and facilitates viral entry. Proc Natl Acad Sci U S A. 2008 Jun 3;105(22):7809-14. Epub 2008 May 19. PMID:18490652 doi:10.1073/pnas.0711241105
- ↑ Van Eerdewegh P, Little RD, Dupuis J, Del Mastro RG, Falls K, Simon J, Torrey D, Pandit S, McKenny J, Braunschweiger K, Walsh A, Liu Z, Hayward B, Folz C, Manning SP, Bawa A, Saracino L, Thackston M, Benchekroun Y, Capparell N, Wang M, Adair R, Feng Y, Dubois J, FitzGerald MG, Huang H, Gibson R, Allen KM, Pedan A, Danzig MR, Umland SP, Egan RW, Cuss FM, Rorke S, Clough JB, Holloway JW, Holgate ST, Keith TP. Association of the ADAM33 gene with asthma and bronchial hyperresponsiveness. Nature. 2002 Jul 25;418(6896):426-30. Epub 2002 Jul 10. PMID:12110844 doi:10.1038/nature00878
- ↑ Wu E, Croucher PI, McKie N. Expression of members of the novel membrane linked metalloproteinase family ADAM in cells derived from a range of haematological malignancies. Biochem Biophys Res Commun. 1997 Jun 18;235(2):437-42. PMID:9199213 doi:10.1006/bbrc.1997.6714
- ↑ Muniz JR, Ambrosio AL, Selistre-de-Araujo HS, Cominetti MR, Moura-da-Silva AM, Oliva G, Garratt RC, Souza DH. The three-dimensional structure of bothropasin, the main hemorrhagic factor from Bothrops jararaca venom: insights for a new classification of snake venom metalloprotease subgroups. Toxicon. 2008 Dec 1;52(7):807-16. Epub 2008 Sep 27. PMID:18831982 doi:10.1016/j.toxicon.2008.08.021
- ↑ 14.0 14.1 14.2 14.3 14.4 Takeda S, Igarashi T, Mori H, Araki S. Crystal structures of VAP1 reveal ADAMs' MDC domain architecture and its unique C-shaped scaffold. EMBO J. 2006 Jun 7;25(11):2388-96. Epub 2006 May 11. PMID:16688218 Cite error: Invalid
<ref>tag; name "Takeda" defined multiple times with different content - ↑ Zhu Z, Gao Y, Zhu Z, Yu Y, Zhang X, Zang J, Teng M, Niu L. Structural basis of the autolysis of AaHIV suggests a novel target recognizing model for ADAM/reprolysin family proteins. Biochem Biophys Res Commun. 2009 Aug 14;386(1):159-64. Epub 2009 Jun 6. PMID:19505434 doi:10.1016/j.bbrc.2009.06.004
- ↑ Gomis-Ruth FX. Structural aspects of the metzincin clan of metalloendopeptidases. Mol Biotechnol. 2003 Jun;24(2):157-202. PMID:12746556 doi:10.1385/MB:24:2:157
- ↑ Odell GV, Ferry PC, Vick LM, Fenton AW, Decker LS, Cowell RL, Ownby CL, Gutierrez JM. Citrate inhibition of snake venom proteases. Toxicon. 1998 Dec;36(12):1801-6. PMID:9839664
- ↑ Marques-Porto R, Lebrun I, Pimenta DC. Self-proteolysis regulation in the Bothrops jararaca venom: the metallopeptidases and their intrinsic peptidic inhibitor. Comp Biochem Physiol C Toxicol Pharmacol. 2008 May;147(4):424-33. Epub, 2008 Feb 5. PMID:18325841 doi:10.1016/j.cbpc.2008.01.011